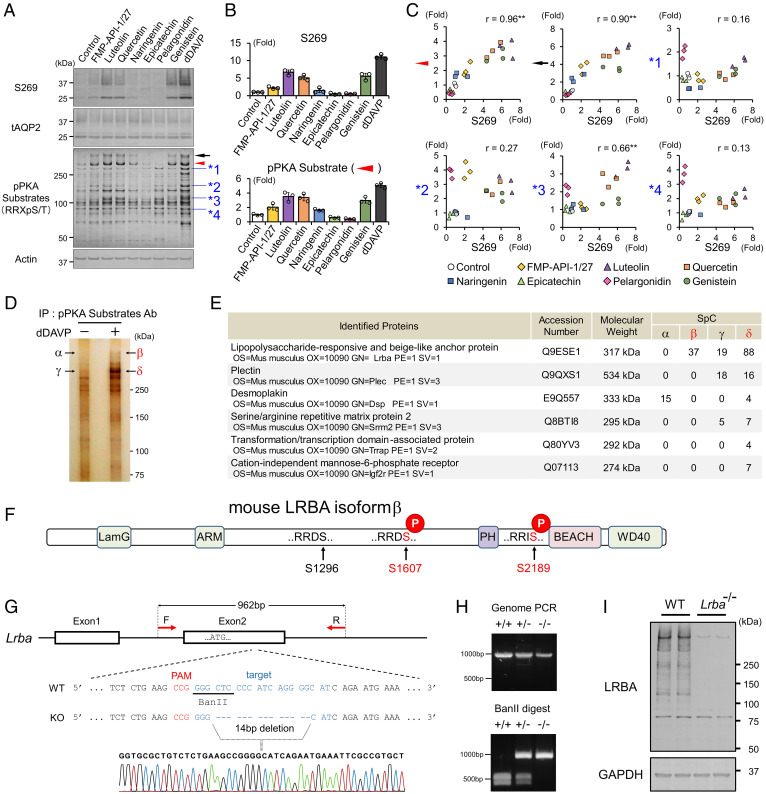
Fig. 1.
FMP-API-1/27 and flavonoids phosphorylate LRBA and AQP2 in a correlated manner. (A–C) Screening for PKA substrates whose phosphorylation are well correlated with pAQP2-S269. (A) Representative blots of pAQP2-S269 and pPKA substrates. (B) Phosphorylation levels of AQP2-S269 and the PKA substrate (indicated by red arrowhead) are quantified by densitometric analysis (n = 3). (C) Phosphorylation levels of AQP2-S269 and PKA substrates (indicated by arrowheads) are strongly correlated in scatterplots. (D and E) Immunoprecipitated PKA substrates (indicated by β and δ) are identified as LRBA by liquid chromatography–tandem mass spectrometry (LC-MS/MS). (D) Representative silver staining of pPKA substrates immunoprecipitated by pPKA substrate antibody. (E) Identified proteins by LC-MS/MS. (F) PTM analysis reveals the RRXS sites of LRBA phosphorylated by dDAVP. (G and H) Lrba knockout mice are generated by CRISPR/Cas9 genome-editing technology. (G) The target sequence for Lrba gene editing. (H) Genotyping of Lrba knockout mice after BanII digestion of the PCR products from genomic DNA. (I) Anti-LRBA antibody detects renal LRBA in WT mice. (C) Pearson correlation coefficient r value and two-sided Student's t test. **P < 0.01. Ab, antibody; IP, immunoprecipitation; PAM, protospacer adjacent motif; SpC, spectral count.