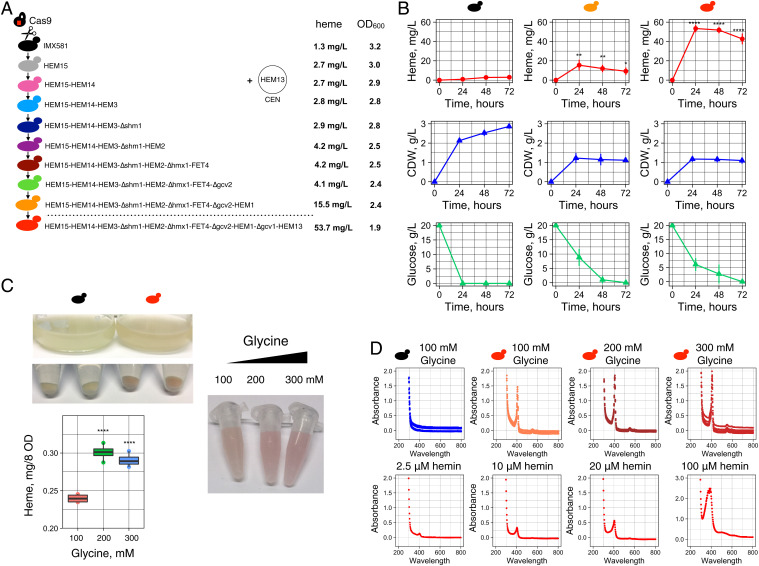
Fig. 4.
CRISPR-Cas9 genome engineering for increased heme production. (A) The IMX581 strain carrying CRISPR-Cas9 gene integrated in the genome was used to carry the combinatorial engineering of heme gene targets deduced by Yeast8 and ecYeast8 genome-scale model. The gene integrations and deletions were performed using the gRNA constructs targeting different genome loci. The gene HEM13 was overexpressed from the centromeric plasmid. The HEM13 expression cassette was integrated into the genome in the final strain. Absolute heme (mg/L) was extracted from the entire biomass of the strains. (B) Heme production, CDW, and glucose consumption in different strains at 24, 48, and 72 h of cultivation in buffered SD ura- or SD with 2% glucose, 100 mM glycine supplemented with 0.1 mM Fe3+. Four biological replicates (transformants) were used in the experiment. Error bars represent the SD. Commercial hemin was used to calibrate data. Strains: IMX581 carrying an empty vector; IMX581/HEM15 HEM14 HEM3 Δshm1 HEM2 Δhmx1 FET4 Δgcv2 HEM1 carrying the HEM13 centromeric plasmid; IMX581/HEM15 HEM14 HEM3 Δshm1 HEM2 Δhmx1 FET4 Δgcv2 HEM1 Δgcv1 carrying HEM13 expression cassette integrated into genome. Statistical analysis was performed using one-way ANOVA (*P ≤ 0.02741, **P ≤ 0.00594, ****P ≤ 0). (C) The culture, cells, and cell extracts (obtained with oxalic acid treatment) of engineered strain IMX581/HEM15 HEM14 HEM3 Δshm1 HEM2 Δhmx1 FET4 Δgcv2 HEM1 Δgcv1 HEM13 had a red color. Increasing the glycine amount from 100 to 300 mM resulted in a further increase in heme production. Statistical analysis was performed using one-way ANOVA (****P ≤ 0.00007). (D) Spectral analysis of yeast extracts (obtained with oxalic acid treatment) shows the presence of the Soret peak (at 400 nm) characteristic to heme in IMX581/HEM15 HEM14 HEM3 Δshm1 HEM2 Δhmx1 FET4 Δgcv2 HEM1 Δgcv1 HEM13 strain. Hemin (2.5, 10, 20, and 100 μM) spectra were used in comparison.