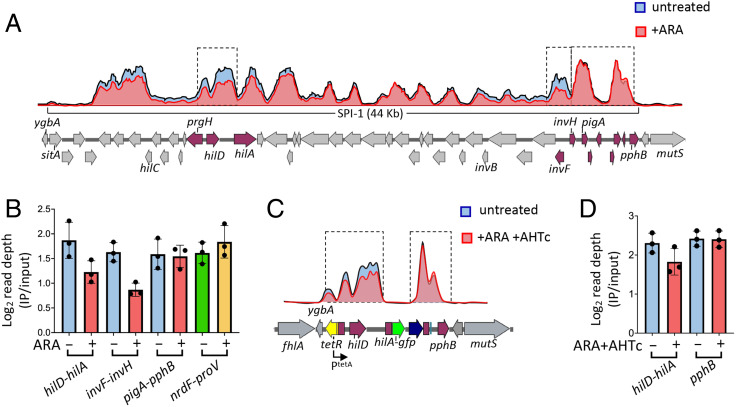
Fig. 5.
NusG depletion affects H-NS binding to specific portions of SPI-1. (A) Representative ChIP-Seq profiles from NusG-depletable strain MA13748 (hns-3xFLAG) grown to early stationary phase in the presence or absence of 0.1% ARA. (B) Read depth quantification in the sections framed by the dashed rectangles in A and in the nrdF-proV intergenic region. Read depth values (determined by the bedcov tool of the Samtools suite) were normalized to the values from the entire genome. The results shown represent the ratios between the normalized values from IP samples and those from input DNA. (C) Representative ChIP-Seq profiles from NusG-depletable strain MA14513 (Δ[sitA-prgH]::tetR-PtetA, hilA::gfpSFΔK28, hns-3xFLAG) grown to early stationary phase in the presence or absence of ARA + AHTc. (D) Read depth quantification in the intervals framed by the dashed rectangles in C. Read depth was calculated and normalized as in B. The data in B and C represent the means from three independent ChIP-Seq experiments (with error bars indicating SDs).