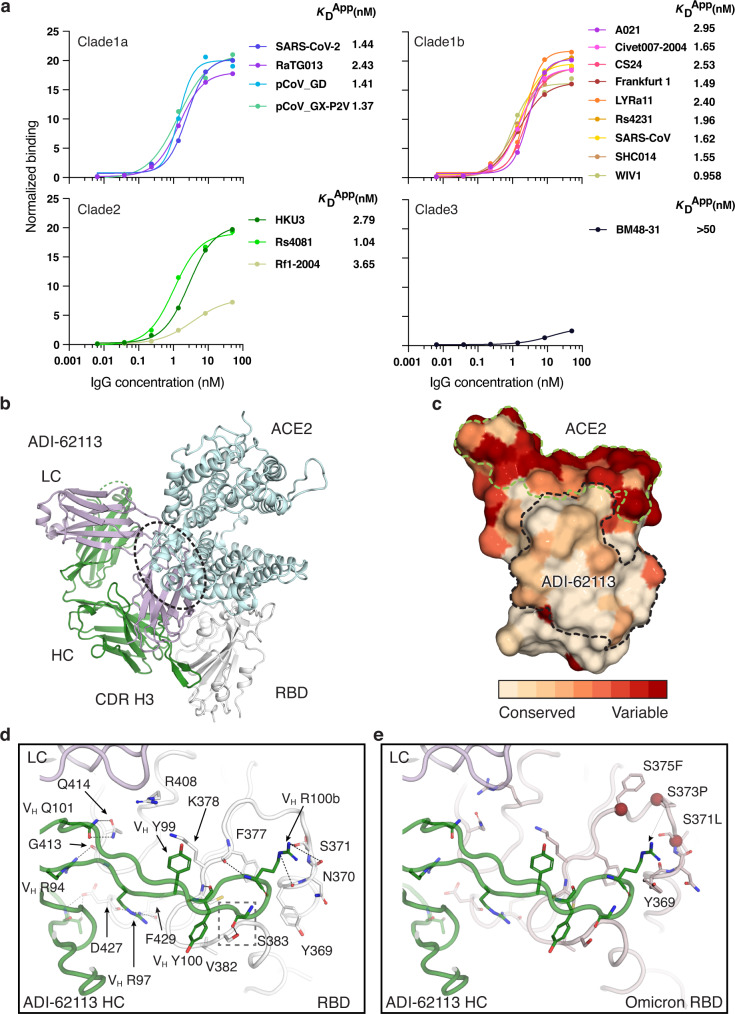
Fig. 1. ADI-62113 binds a highly conserved site on SARS-CoV-2 RBD and cross-reacts with many sarbecoviruses.
a ADI-62113 shows a broad spectrum of cross-reactivity to sarbecoviruses. RBDs from viruses in clade 1a (SARS-CoV-2-like viruses), clade1b (SARS-CoV-like viruses), clade 2, and clade 3 were displayed on the surface of yeast for binding kinetics analysis. Clade 1a and 1b viruses use ACE2 as an entry receptor. Clade 2 and 3 viruses do not bind ACE2 but contain homologous sequences to SARS-CoV-2. b Composite structure showing ADI-62113 competition with ACE2 binding for SARS-CoV-2 RBD. ADI-62113 Fab, ACE2, and SARS-CoV-2 RBD are shown in a ribbon representation. ACE2 is superimposed on the crystal structure of ADI-62113 with SARS-CoV-2 RBD based on PDB ID: 6M0J. White, RBD; pale cyan, ACE2; dark green, heavy chain; lavender, light chain. c ADI-62113 binds a highly conserved surface, while the receptor binding site (RBS) is highly variable across sarbecoviruses. SARS-CoV-2 RBD is shown in surface representation and colored by conservation across 169 sarbecovirus sequences (Supplementary Data 1). Dashed lines show the outline of the buried surface area (BSA) on the RBD by the human receptor ACE2 (green) and ADI-62113 (black). 81% of the ADI-62113 epitope surface is buried by the heavy chain and 19% by the light chain. CDR H3 contributes 69% of the total BSA. d CDR H3 interacts with highly conserved residues in the RBD. The crystal structure of ADI-62113 in complex with SARS-CoV-2 RBD is shown in tube representation. Residues involved in the interface between ADI-62113 and SARS-CoV-2 RBD are shown in sticks. Dashed lines represent hydrogen bonds or salt bridges. VH R94, R97, R100b, G100d, and N101 hydrogen bond with the RBD. VH Y99, Y100, and R100b have hydrophobic and amide-π interactions with the RBD. The dashed box indicates potential space constraints for the glycine in the YYDRxG motif in CDR H3 as shown in Fig. 2d. e The epitope site of ADI-62113 is much less impacted by mutations in VOCs such as Omicron BA.1. Epitope and paratope residues are shown in sticks with no labels (for labels, please see d). Mutation sites in Omicron are shown as red spheres with side chains as sticks and labeled with corresponding residue changes. A composite structure of ADI-62113+Omicron RBD was formed by superimposition of structures of ADI-62113+wildtype RBD (this study) and the Omicron RBD + ACE2 (PDB ID: 7T9L). The same perspective view as in d was used for parallel comparison. Interactions between ADI-62113 and RBDs are largely retained between wildtype (d) and Omicron RBD (e).