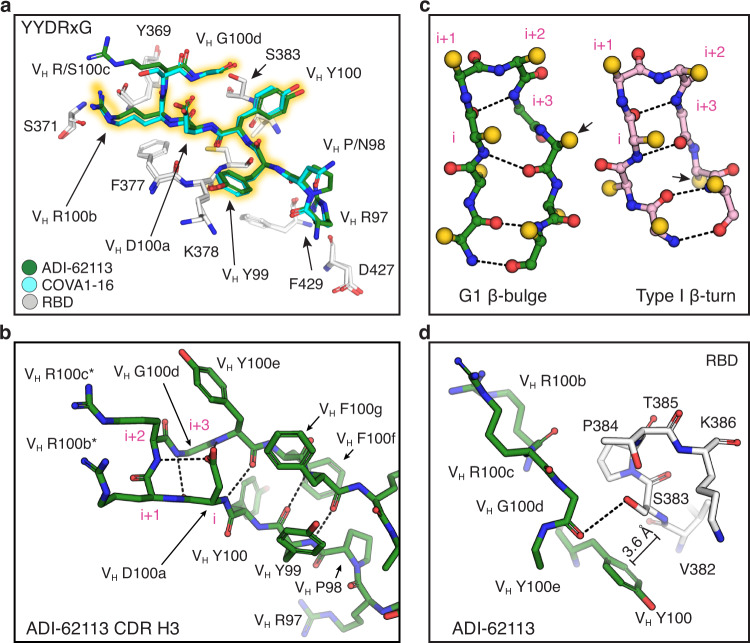
Fig. 2. The YYDRxG motif is a recurring feature in CDR H3 for RBD binding.
Hydrogen bonds are shown as dashed lines. Key epitope and paratope residues are shown in sticks and labeled. a Comparison of CDR H3 interactions of ADI-62113 and COVA1-16 with the RBD. White, SARS-CoV-2 RBD; green, ADI-62113; cyan, COVA1-16. The 99YYDRxG100d hexapeptide is highlighted in yellow to show its conserved structure in CDR H3 that interacts with the RBD. b The YYDRxG motif is located at the tip of CDR H3 and precedes a G1 β-bulge in the descending strand of the hairpin structure. Residues in the β-turn at the tip of CDR H3 are numbered i to i + 3 (magenta). The VH D100a carboxyl (residue i) hydrogen bonds to backbone amide of VH R100c (i + 2) in the β-turn as well as the backbone amide of VH Y100e in the β-bulge. Asterisk (*) indicates somatically mutated residue. c Backbone comparison of an inserted β-bulge versus a standard β-strand in a β-sheet. Schematic backbones show the β-harpin in ADI-62113 CDR H3 that contains a G1 β-bulge following the glycine residue (i + 3) in the β-turn and comparison with a standard β-hairpin also with a type 1 β-turn at its tip (PDB ID: 4H5U). Amino-acid side chains are simplified as gold spheres. Arrows indicate the register change between the two motifs due to an additional residue in the β-hairpin in ADI-62113. d Limited space between residue VH G100d and RBD favors a glycine residue at this position in the β-bulge. A hydrogen bond is formed between the VH G100d carbonyl oxygen and S383 hydroxyl in the RBD. An amide-π interaction is formed between VH Y100 and the peptide backbone of 382VS383 of SARS-CoV-2 RBD.