Fig. 5. T2T-CHM13 improves clinical genomics variant calling.
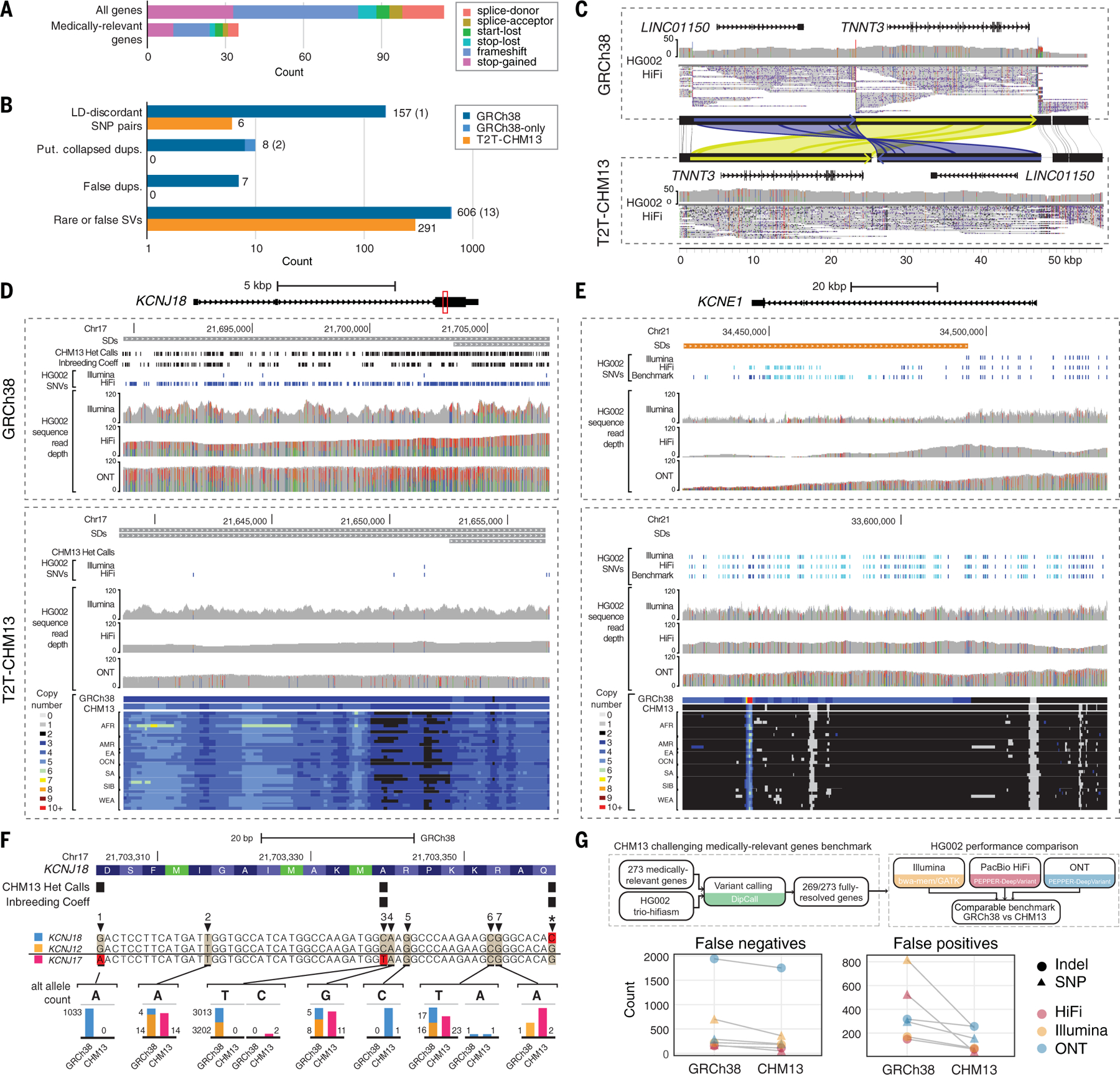
(A) Numbers of potential loss-of-function mutations in the T2T-CHM13 reference. (B) The counts of medically relevant genes affected by genomic features and variation in GRCh38 (blue) and T2T-CHM13 (orange) are depicted as bar plots on logarithmic scale. Light blue indicates genes affected in GRCh38 where homologous genes were not identified in T2T-CHM13 due to inability to lift over, with counts included in parentheses. (C) An example erroneous GRCh38 complex SV corrected in T2T-CHM13 affecting TNNT3 and LINC01150, displayed by sequence comparison using miropeats (88) with homologous regions colored in green and blue, respectively. HG002 PacBio HiFi data are displayed showing read coverages and mappings from IGV, with allele fractions of variant sites colored (red, T; green, A; blue, C; black, G) within histograms of read depth (0 to 50). (D and E) Snapshots of regions using IGV and UCSC Genome Browser representing a collapsed duplication in GRCh38 corrected in T2T-CHM13 affecting KCNJ18 (D) and a false duplication in GRCh38 affecting most of KCNE1 (E). SDs depicted on top are colored by sequence similarity to paralog (gray, 90 to 98%; orange, >99%). Read mappings and variants from HG002 Illumina, PacBio HiFi, and ONT (mappings only), with homozygous (light blue) and heterozygous (dark blue) variants depicted as dashes. Colors within histograms of read depth (0–120) are the same as described in (C). Copy number estimates, displayed as colors indicated in legends, across k-merized versions of the GRCh38 and T2T-CHM13 references as well as representative examples of the SGDP individuals. (F) An example CDS region of KCNJ18 (highlighted as a red box in D), with amino acids colored in alternating shades of blue and potential start codons (methionines) labeled in green using the UCSC Genome Browser codon-coloring scheme. Alignments of KCNJ18 (blue), KCNJ12 (orange), and KCNJ17 (pink) along with allele counts of variants in each gene identified on GRCh38 and T2T-CHM13 are shown as bar plots (to approximate scale per variant), with examples 1 to 7 described in table S14. (G) Schematic depicts a benchmark for 269 challenging medically relevant genes for HG002. The number of variant-calling errors from three sequencing technologies on each reference is plotted.
