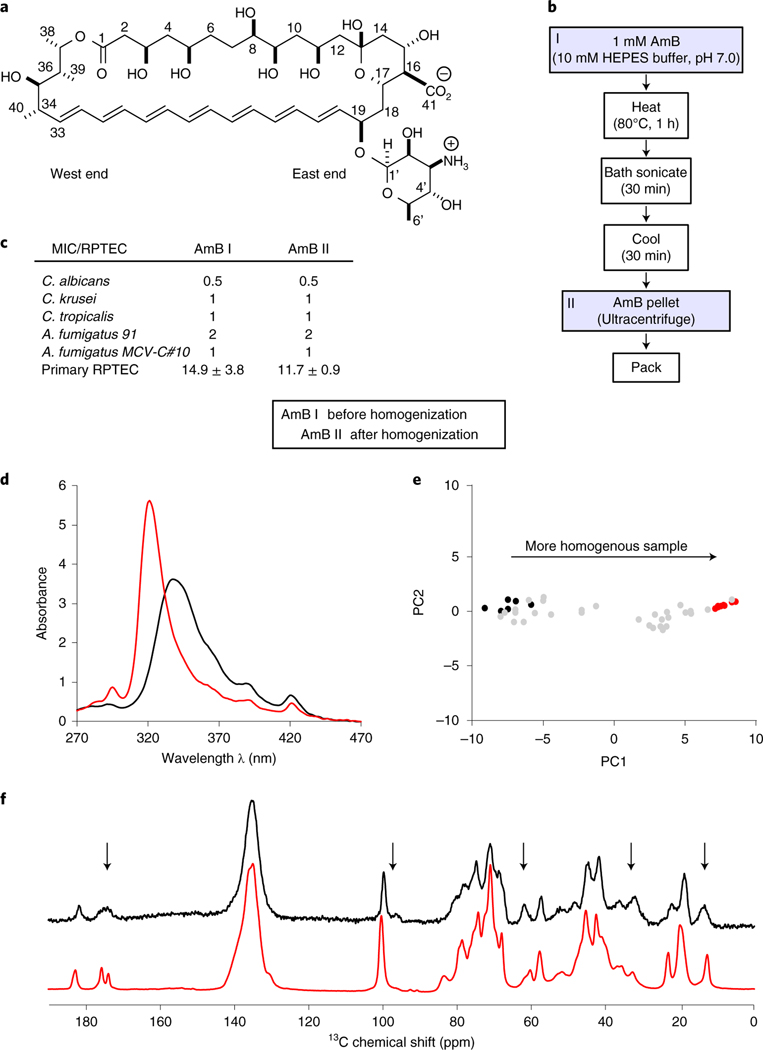
Fig. 1 |. An improved AmB sponge preparation technique that reproducibly yields homogeneous SSNMR samples.
a, Molecular structure of AmB polyene macrolide. b, Schematic representation of the process for obtaining homogenous AmB assemblies for SSNMR spectroscopy experiments. First, 1 mM AmB aggregate in 10 mM HEPES buffer pH 7.0 is prepared. This sample is then heated at 80 °C for 1 h, followed by bath sonication for 30 min and cooling at room temperature for another 30 min. Next, the sample is pelleted in the ultracentrifuge and spectrophotometrically analyzed before packing it into the SSNMR rotor. c, Biological analysis (MIC and MTC) of the AmB aggregates before (AmB I) and after (AmB II) homogenization. MIC against yeast pathogens (C. albicans, C. krusei, C. tropicalis and A. fumigatus) and MTC values causing 90% toxicity against human primary RPTECs ± percent error standard deviation. MIC and MTC values are given in μM. d, UV–Vis spectra of AmB aggregates before (black) and after (red) homogenization. e, Sample quality validation by PCA of the UV spectrum. PC1/PC2 projection of the heterogeneous (black) and homogenous (red) samples that were obtained after homogenization. f, 13C 1D SSNMR spectra (with 50 Hz line broadening) of the [U-13C]AmB before (black) and after (red) homogenization. Black arrows represent carbon sites with changes in resolution.