Figure 3. Conserved binding motifs on the catalytic domain surface.
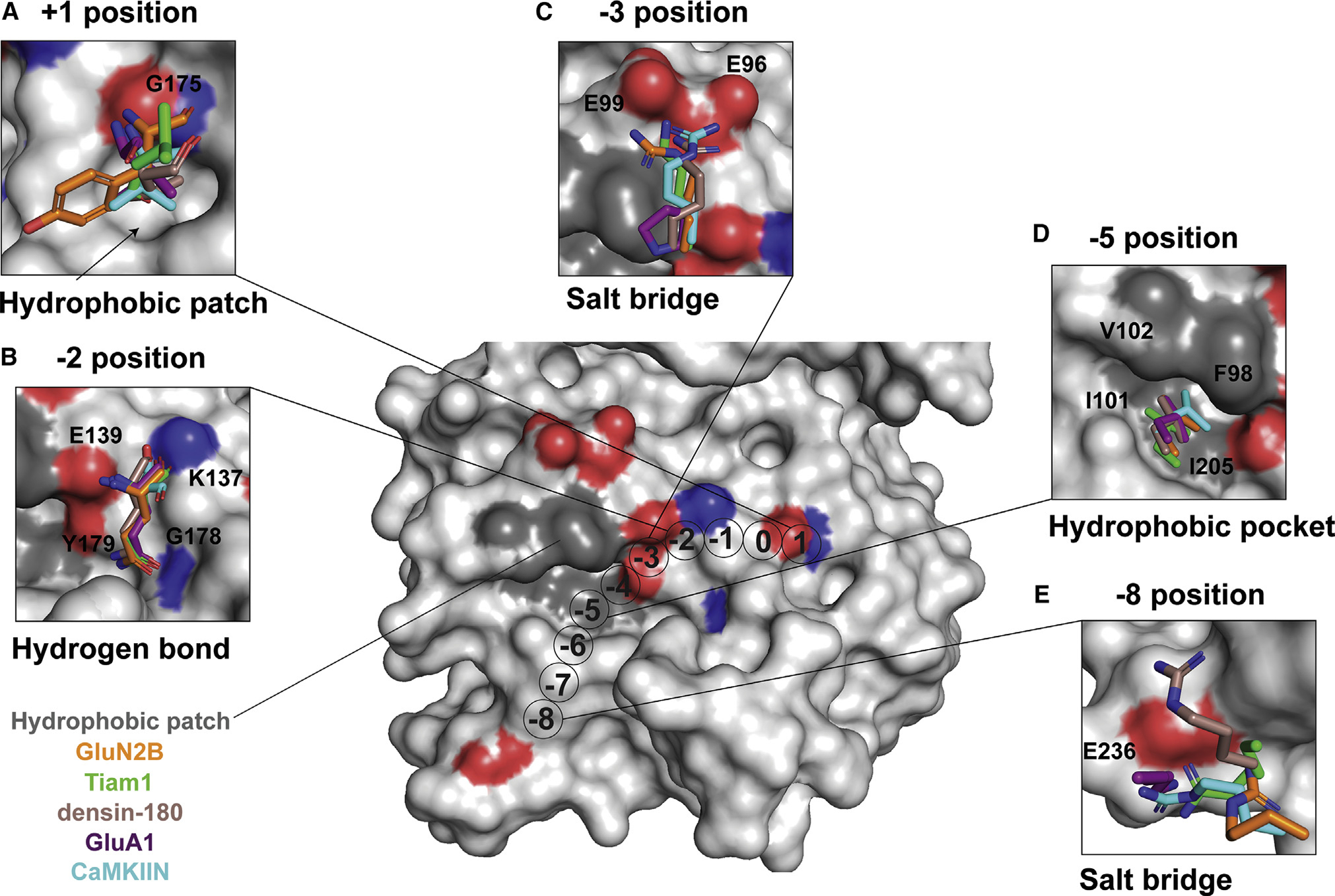
Shown is a surface representation of the CaMKII kinase domain, highlighting residues that mediate interactions with binding partners. The color code matches previous figures (GluN2B, orange; Tiam1, green; densin-180, brown; GluA1, purple; CaMKIIN, cyan).
(A) At the +1 position, a hydrophobic patch (arrow) is formed by F173, P177, L185, and Y222. GluA1, CaMKIIN, and densin-180 have Val or Ile at the +1 position, which are buried in this hydrophobic groove. Backbone atoms of the +1 residue hydrogen bond with the backbone of G175 (highlighted red and blue on the structure).
(B) At the −2 position, all interactors have a glutamine except for CaMKIIN, which has a serine. The glutamine side-chain amide oxygen forms a hydrogen bond with the backbone of G178, and the amino group interacts with the side chain of Y179. The backbone carbonyl interacts with the side chain of K137, and the backbone amino group interacts with E139.
(C) At the −3 position, lysine or arginine interacts with E96 and E99. The basic residues of interaction partners are positioned 2.4–4.2 Å between E96 and E99. GluA1 has a proline at the −3 position, which is flipped away from E96/99.
(D) At the −5 position, a conserved leucine across all interactors nestles into a hydrophobic pocket formed by F98, I101, V102, and I205.
(E) Lysine or arginine at the −8 position forms a salt bridge with E236.
