Figure 7. GluN2B binding interferes with autoinhibition.
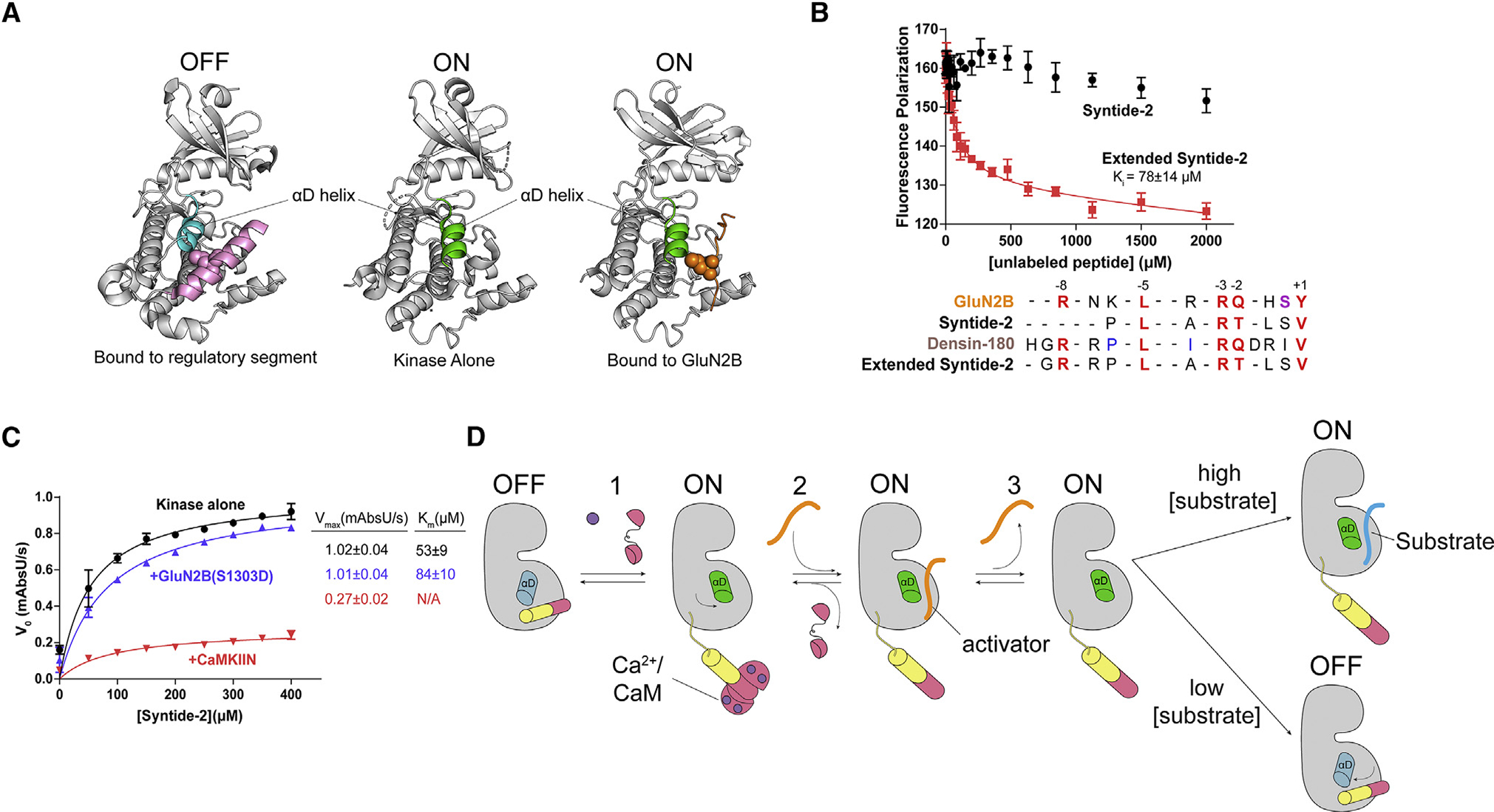
(A) Comparison of the αD helix between crystal structures of the autoinhibited with regulatory segment (pink) bound (left; PDB: 2VZ6), uninhibited kinase with no regulatory segment (center; PDB: 6VZK), and active kinase with GluN2B (orange) bound (right; PDB: 7UIS). The αD helix goes from rotated inward in the off-state (blue) to rotated outward 45° in the on state (green).
(B) Competition assay against GluN2B using Syntide-2 (black) and the extended version of Syntide-2 (red). Error bars indicate standard deviation from triplicate at each data point. Sequence alignment of GluN2B, Syntide-2, densin-180, and the extended Syntide-2 is shown below the graph. For clarity, the alignments start at the +1 position (C-terminal end) and contain the −8 basic residue (N-terminal end). Full sequences used are listed in the STAR Methods.
(C) Coupled kinase assay results with kinase alone and in the presence of GluN2B and CaMKIIN. The significance of the change in Km values is significant when using unpaired t test with Welch’s correction (p = 0.0208).
(D) Proposed model of maintaining CaMKII activity by binding to a high-affinity activator. CaMKII binds Ca2+/CaM and is activated, and the αD helix rotates out (Rxn 1). A high-affinity activator binds to the substrate binding site (Rxn 2). When the Ca2+ signal dissipates, CaM dissociates from CaMKII, but the activator remains bound, competing with the regulatory segment. The activator dissociates, but the αD helix remains in the active conformation (Rxn 3). In high [substrate], the substrate will bind and be phosphorylated. In low [substrate], the activator will rebind or the αD helix will rotate back in to accommodate regulatory segment binding.
