Fig. 2 |. Pre-analytical factors impacting the accuracy of cfDNA analysis.
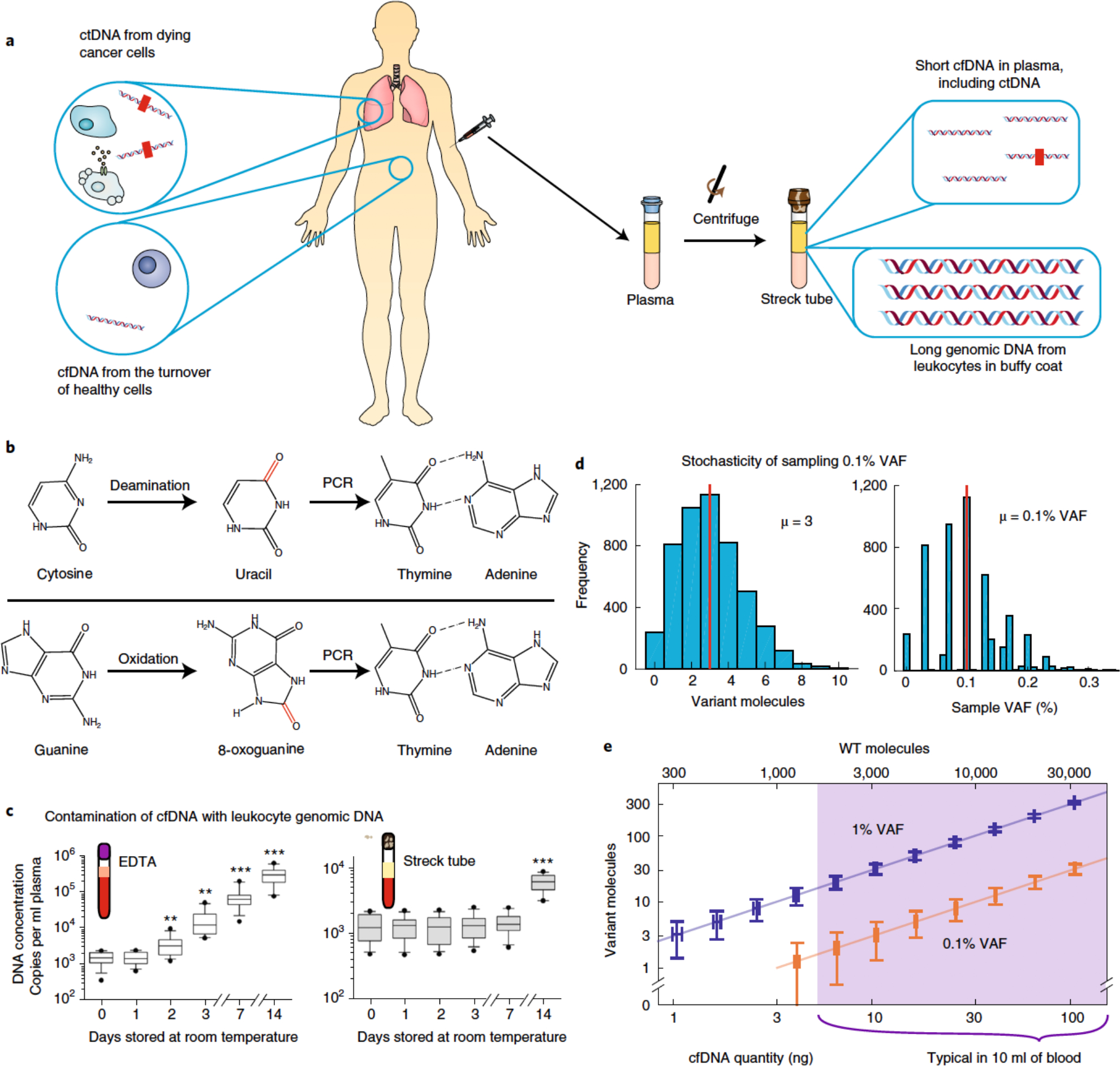
a, Whereas the buffy-coat layer of blood is rich in genomic DNA from peripheral mononuclear blood cells, blood plasma contains relatively low quantities of extracellular DNA (that is, cfDNA). cfDNA results from dying cells in the entire body, including healthy cells (depicted in white) and tumour cells (shown in brown) dying from apoptosis, necrosis and immune cytotoxicity. Thus, only a small fraction of cfDNA comprises tumour-derived ctDNA. The red rectangles within the ctDNA strands denote tumour-specific mutations. b, cfDNA in blood may be damaged during sample collection, transport and storage, resulting in modified nucleosides that are incorrectly recognized by DNA polymerases during PCR amplification. This leads to amplicon DNA sequences with variants that may be interpreted as cancer-specific mutations. The schematics show cytosine deamination and guanine oxidation, the two most commonly observed types of DNA damage. c, Contamination of cfDNA with genomic DNA from leukocytes. Except for blood cancers, genomic DNA from leukocytes will not contain cancer-specific ctDNA. Thus, contamination of cfDNA with leukocyte genomic DNA will dilute the fraction of cfDNA that contains useful information, rendering the mutation analysis of downstream DNA more difficult and the quantitation of VAF less accurate. EDTA, ethylenediaminetetraacetic acid. d, Poisson distribution of tumour-mutation molecules and VAF in a blood sample. An adult human has roughly 5 l of blood in circulation, and sampling 10 ml of blood for cfDNA analysis introduces variations in VAF owing to small-number statistics. Assuming a ‘ground truth’ of 0.1% cancer-mutation VAF in the entire 5 l of blood supply and a 10 ng sample of cfDNA in a 4 ml plasma sample, the number of cancer-mutation molecules present will range between 0 and 10, corresponding to an observed VAF range of 0–0.3% for any given DNA locus. No technology improvements can transcend this sampling variation; only the use of larger-volume blood samples can mitigate this VAF-irreproducibility challenge. The Matlab code used to generate these results is provided as Supplementary Information. e, Visualization of molecule-number variations owing to cfDNA sampling. The vertical and horizontal error bars show the analytically calculated standard-deviation values for different cfDNA input quantities and mutation VAFs. WT, wild-type. Panel c adapted from ref. 71.
