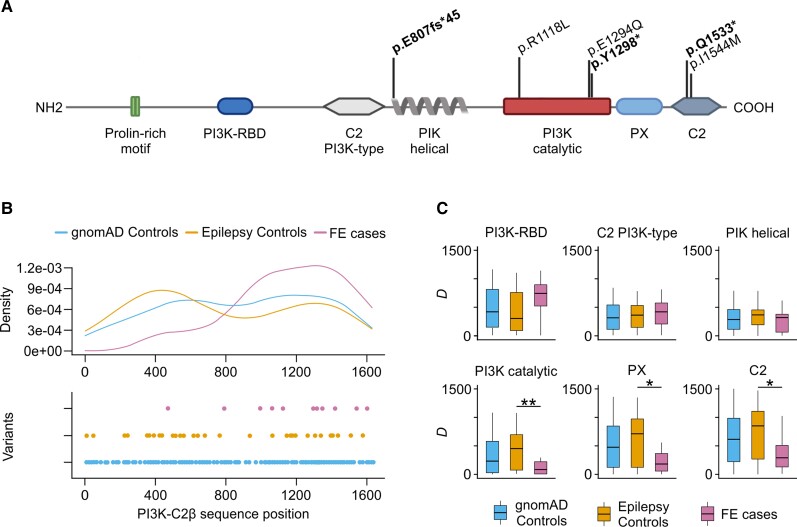
Figure 1.
URVs in PIK3C2B are loss-of-function. (A) Protein-altering, deleterious URVs in PIK3C2B identified in FE patients from this study, and their location with respect to protein length and protein domains according to the Uniprot 000750 record. ‘PI3K Catalytic’ refers to the ‘PI3K/PI4K’ domain intervals in the Uniprot reference. Loss-of-function URVs are shown in bold. (B) Density (top) and point (bottom) distribution of smURVs with respect to the protein sequence positions. Combined smURVs in FE cases from this study and the Epi25 NAFE group are compared to controls from this study and the Epi25 control group. Skewed distribution of FE-associated smURVs towards the C-terminal end of the protein results in a significant difference of CLUMP scores between cases and controls (P = 0.007). (C) Domain-proximity analysis comparing D, the mean distance of smURVs from protein domain boundaries, in cases and controls. The most significant difference in D between cases and controls is found for the PI3K catalytic domain (P = 0.006), followed by the PX (P = 0.017) and C2 (P = 0.024) C-terminal domains. For all analyses, singleton missense variants in gnomAD v2.1.1 controls (allele count ≤1) are also shown.