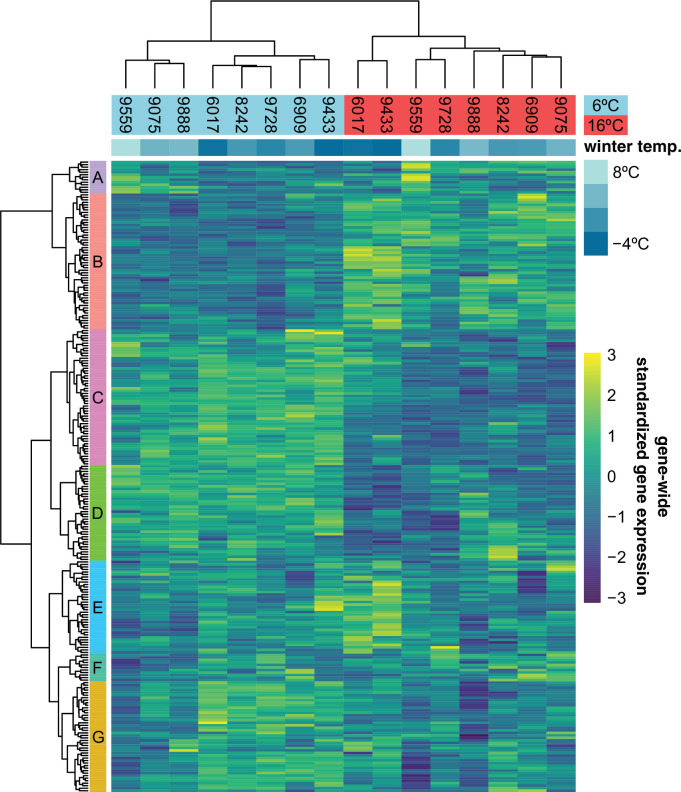
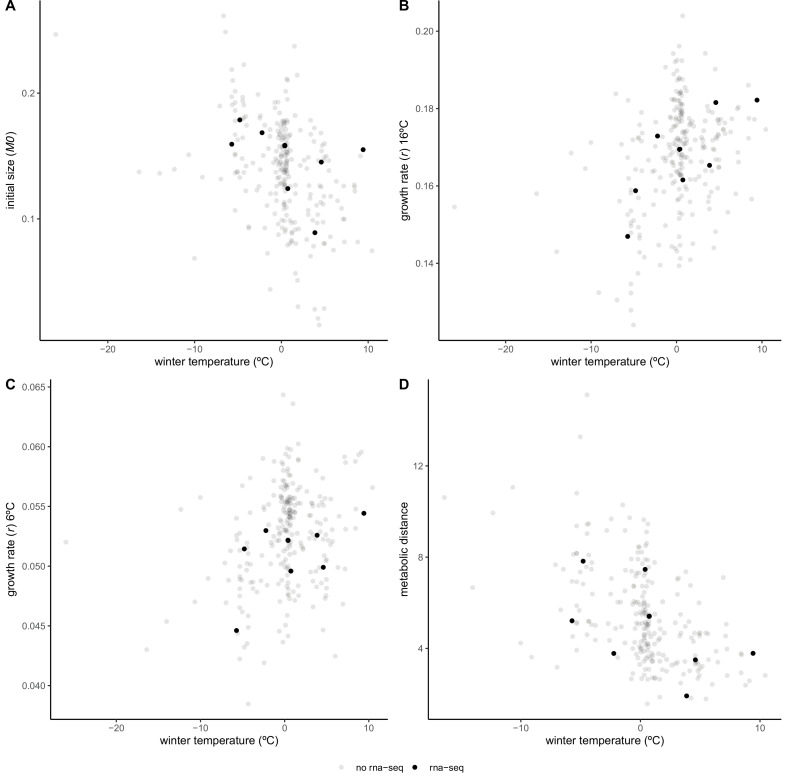
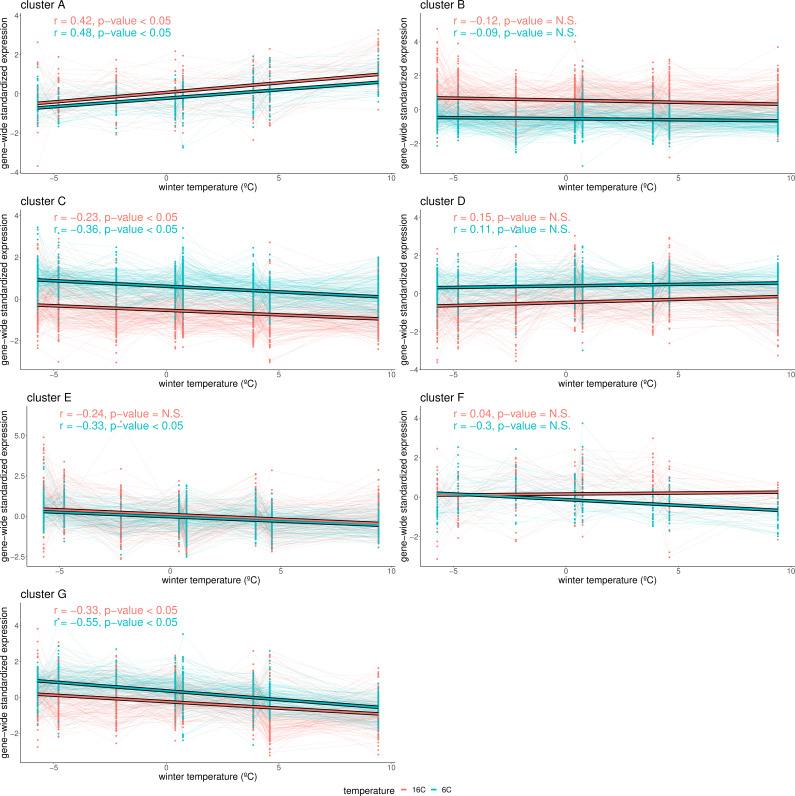
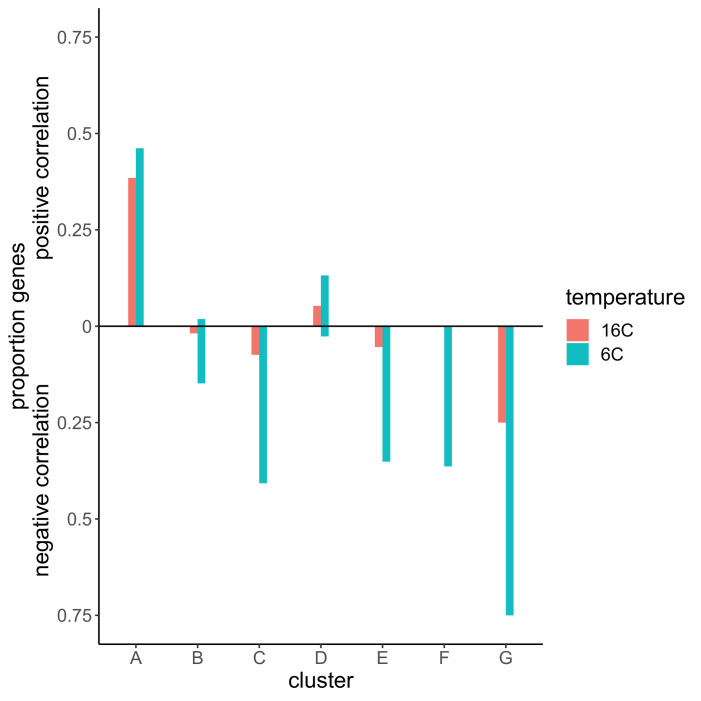
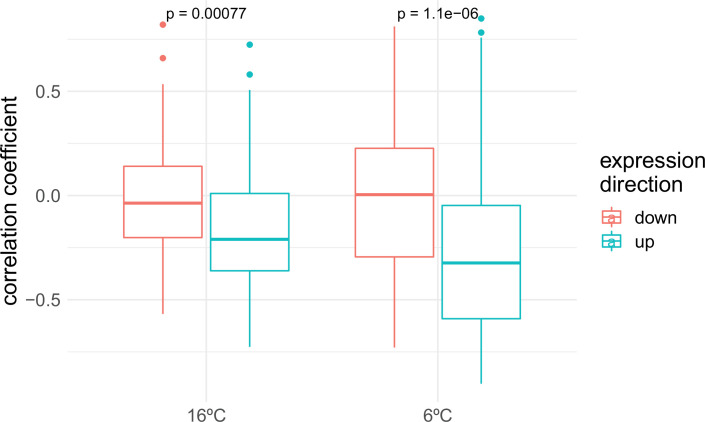
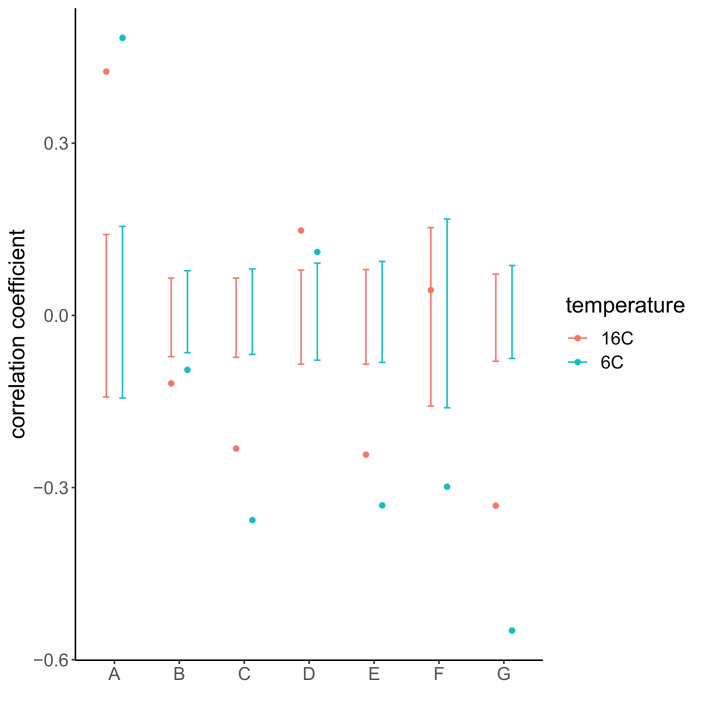
Figure 3. Expression of 251 previously described cold-acclimation genes.
Expression is shown as the gene-wide z-scores of the normalized counts. The z-scores allow for grouping genes with a similar expression behavior over the different accessions in both temperatures. The top bar indicates winter temperature (°C) for each accession’s origin. Both dendrograms along y-axis and x-axis, respectively, show hierarchical clustering of genes, and of accessions in both temperatures.