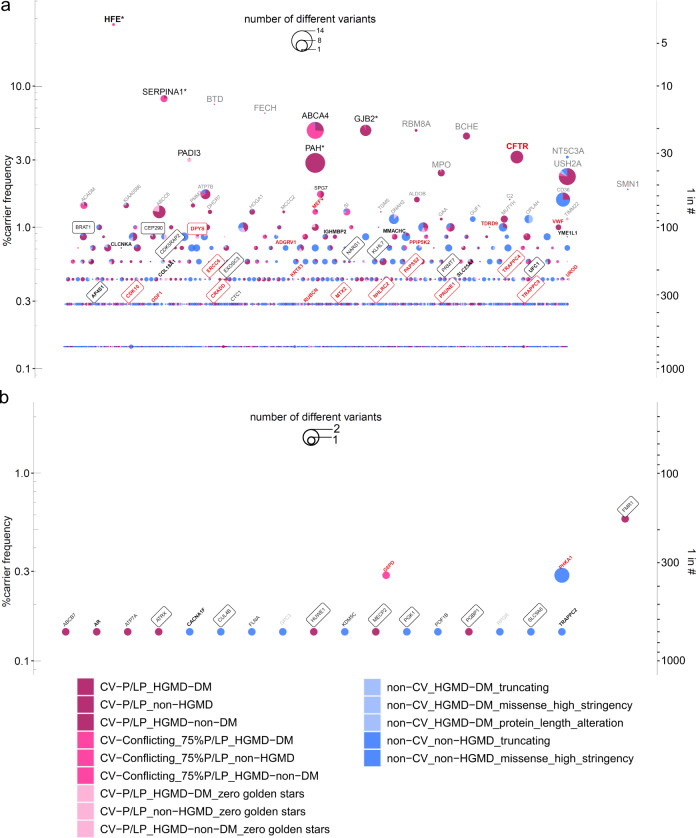
Fig. 2. Carrier frequencies of recessive and X-linked genes observed in the 700 parental samples with indication of at-risk and transmitted at-risk genes and respective variant classification group distributions.
Carrier frequencies of recessive (a) and X-linked genes (b). Carrier frequency was calculated as the percentage of the number of individuals carrying variants of different pathogenicity groups in each gene; on the right y-axis, a descriptive proportion was given. From top to bottom, the genes were sorted according to their carrier frequencies, as well as alphabetically wherever equal. Black letters indicate genes with at least one at-risk parental couple, red for genes with at least one at-risk consanguineous parental couple, bold for genes with at least one at-risk homozygous variant, and boxed for genes with at least one couple having transmitted both at-risk genotypes. Grey letters depict genes with > 1% carrier frequencies that were not found in an at-risk constellation. Stars indicate genes with more than one variant identified in at least one individual. Pie size correlates with the number of different variants in individual genes. The carrier frequencies were calculated among the 700 healthy individuals (350 parental couples) for 3046 recessive genes. The detected variants were shown for 14 variant classification groups according to their levels of pathogenicity, which include ClinVar pathogenic/likely pathogenic (P/LP) (CV-P/LP, in dark pink), ClinVar conflicting with 75% P/LP entries (CV-conflicting_75%P/LP, in pink), both stratified according to disease-causing mutation (DM) classification in the Human Gene Mutation Database (DM, non-HGMD, or HGMD-non-DM). The next groups include ClinVar variants with zero golden stars (in light pink), non-ClinVar with disease-causing mutation classification in the Human Gene Mutation Database (non-CV_HGMD-DM, in light blue) sub-categorized into variant functional classes including truncating, high stringency missense, or protein-length alteration, Non-CV_non-HGMD (blue) variants were sub-categorized according to variant functional classes including truncating, or high stringency missense.