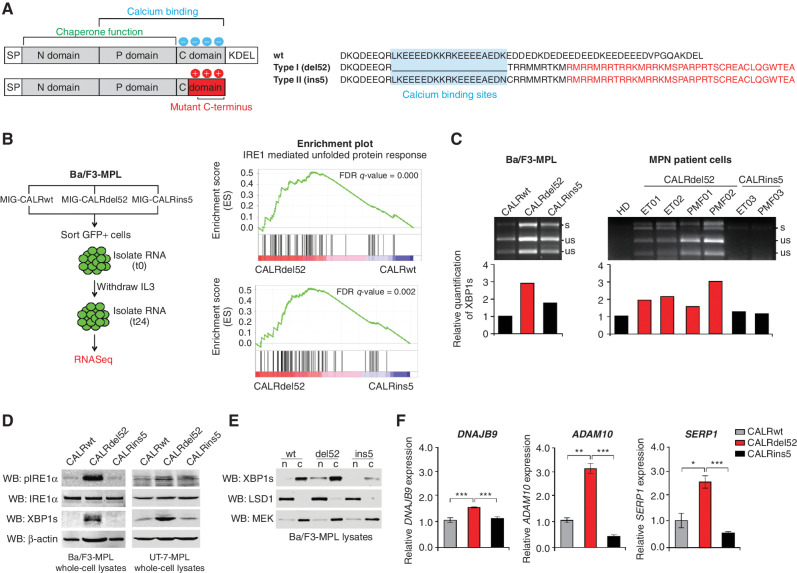
Figure 1.
Type I CALRdel52 mutations differentially activate the IRE1α/XBP1 pathway of the unfolded protein response (UPR). A, Left, structural comparison of wild-type (top) and mutant (bottom) calreticulin (CALR). CALR contains three distinct domains: the N-domain and P-domain are responsible for the chaperone function of the protein, although the P domain also contains one high-affinity Ca2+ binding site; the C-terminal domain binds Ca2+ with a series of acidic amino acids (denoted by blue circles), and terminates at an ER retention signal (KDEL). SP, ER signal peptide. CALR mutations occur in the C-terminal Ca2+ binding domain, and produce an identical 36 amino acid mutant C-terminal tail (red shaded region). The mutant C-terminus is characterized by the replacement of the acidic Ca2+ binding residues with positively charged residues (denoted by red circles), and loss of the KDEL sequence. SP, ER signal peptide. Right, schematic depicting C-terminal amino acid sequence of CALRwt versus type I CALRdel52 and type II CALRins5. CALRins5 retains many of the Ca2+ binding residues present in the wild-type protein, which are lost in the del52-mutant protein (highlighted in blue). 36 amino acid mutant C-terminal tail shared between all CALR-mutant proteins is depicted in red. B, Left, schematic depicting workflow for RNA-sequencing (RNA-seq) experiment in Ba/F3-MPL cells expressing CALR variants in MSCV-IRES-GFP (MIG) backbone. Right, GSEA plots for IRE1α-mediated UPR in Ba/F3-MPL-CALRdel52 cells versus Ba/F3-MPL-CALRwt cells (top) and Ba/F3-MPL-CALRins5 cells (bottom). C, Top, XBP1 splicing assay performed in Ba/F3-MPL cells and MPN patient cells. Top band shows the spliced form of XBP1 (s), bottom bands show the Pst1-digested unspliced form of XBP1 (us). Bottom, quantification of spliced XBP1 band. D, Western blot analysis for phospho-IRE1α, total IRE1α, and spliced XBP1 (XBP1s) in Ba/F3-MPL and UT-7-MPL cells expressing CALR variants. β-Actin was used as a loading control. E, Western blot analysis for XBP1s in CALR-expressing Ba/F3-MPL nuclear and cytosolic extracts. LSD1 was used as a nuclear marker and MEK was used as a cytosolic marker. F, qPCR for XBP1 targets DNAJB9, ADAM10, and SERP1 in UT-7-MPL cells. Each bar represents the average of three replicates. Error bars, SD. Significance was determined by two-tailed Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).