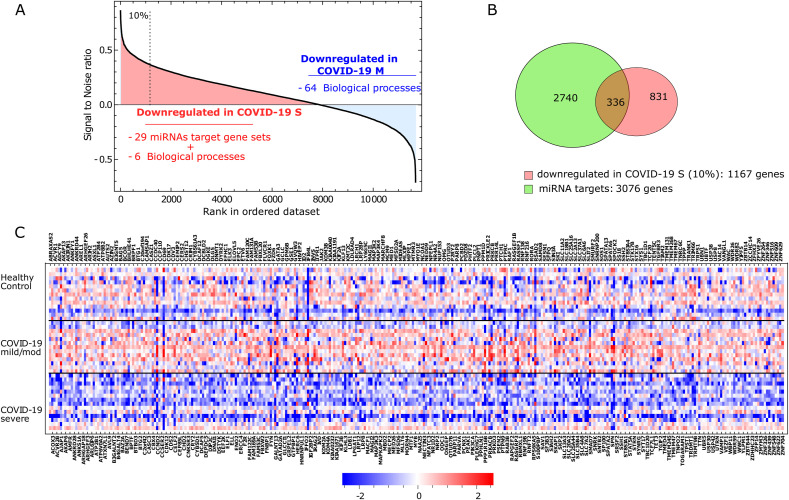
Fig. 5.
The role of epigenetic mechanisms in COVID-19: a regulatory network of miRNAs-target genes
A. Rank of genes according to the Signal2Noise metric as implemented in the GSEA platform (COVID-19 S vs. COVID-19 M) using the media of the normalized counts of the whole cell population in each individual. The top 10% of downregulated genes (rank over the dashed line) was used in the subsequent analysis shown in panels B and C.
B. Venn Diagram showing the overlap between the 3076 gene targets of the 11 miRNAs associated with the target sets enriched in COVID-19 S vs. COVID-19 M and the 1167 top-ranked genes downregulated in COVID-19 S (as explained in panel A). The intersection shows the 336 genes used in panel C.
C. A heat map of the 336 genes that result from the intersection mentioned in panel B. Squares in blue and red represent genes with lower and higher RNA abundance relative to the media of transcript level of all subjects and then log2 transformed, respectively.
COVID-19 S: severe disease; COVID-19 M: mild/moderate disease.