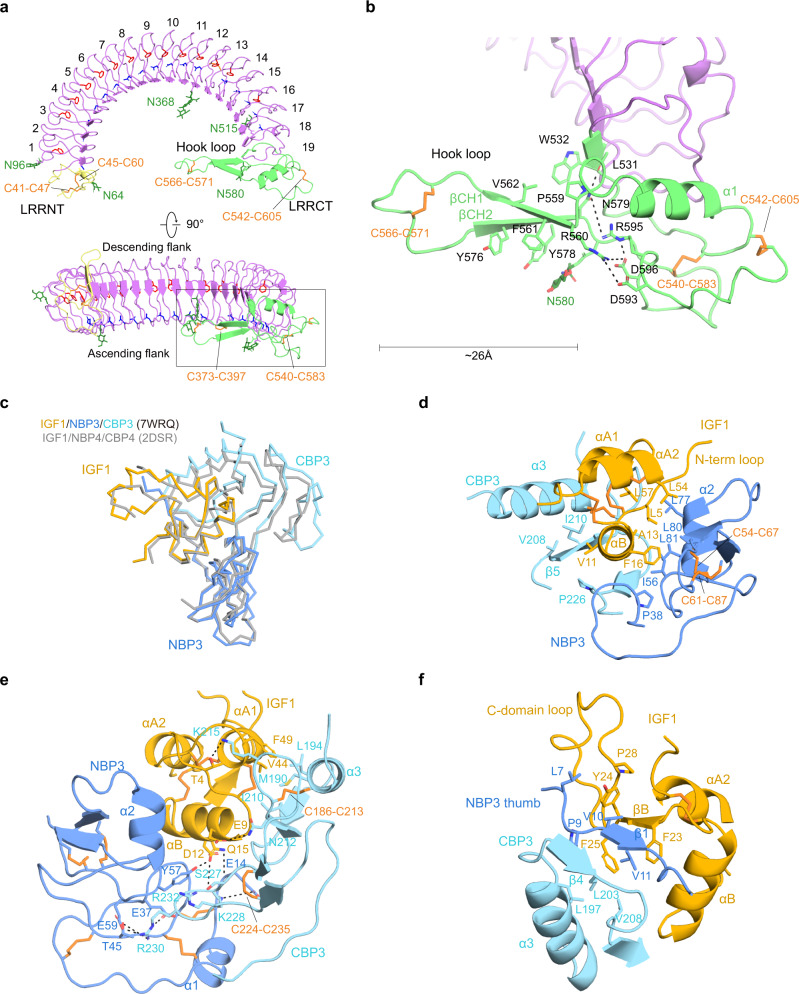
Fig. 2. Structure of ALS and key determinants of IGF1/IGFBP3 binary complex.
a Two different views of the horseshoe-like structure of ALS. LRRNT (yellow), LRR 1–19 (violet), and LRRCT (lime) of ALS are labeled. The conserved asparagine-ladder (blue) and phenylalanine-spine (red) are shown as sticks. Descending and ascending flanks of ALS are labeled (bottom). b Close-up view of the ALS hook loop (boxed in A). Residues involved in stabilization of the ALS hook loop are displayed as sticks and labeled. a, b The glycosylation sites (N64, N96, N368, N515, and N580) and disulfide bridges (C41–C47 and C45–C60 in LRRNT, C373–C397 in LRR motif, C540–C583, C542–C605, and C566–C571 in LRRCT) are indicated. Disulfide bridges (orange) and N-linked glycans (green) are shown as sticks. c Comparison of the IGF1/IGFBP3 in the ternary complex with the structure of IGF1/NBP4/CBP4 complex (gray, PDB: 2DSR). Structures are shown as ribbon diagrams. d–f Close-up views of the interacting interface between the IGF1 B domain ɑ helix and the cleft formed by NBP3 and CBP3 (d), interaction at the interface between IGF1 and the entrance of the cleft, and direct contact between NBP3 and CBP3 (e), and β sheet formed by IGF1, CBP3, and the N-terminal thumb of NBP3 (f). The view for e is a rotation of the view for d ~150° about the y-axis. Key residues involved in each interaction are displayed as sticks and labeled. The ɑ helix (A8-V17) and β strand (G22-Y24) of the IGF1 B domain and the ɑ1 helix (I43-F49) and ɑ2 helix (L54-Y60) of the IGF1 A domain are labeled as ɑB, βB, ɑA1, and ɑA2, respectively.