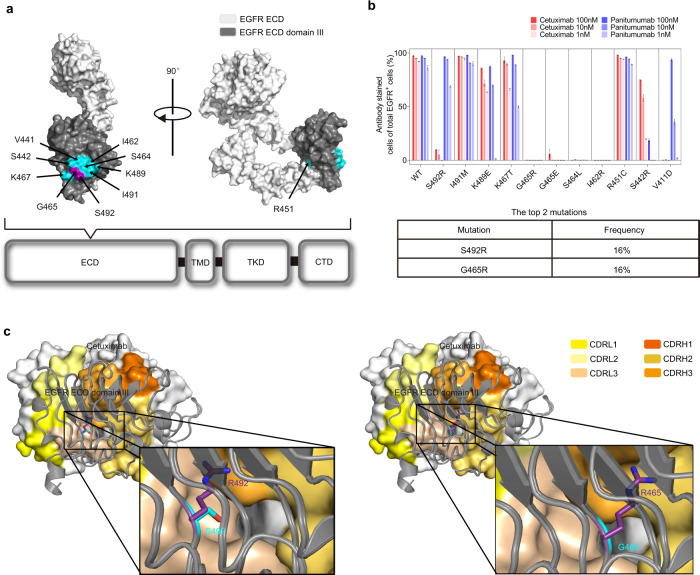
Fig. 1. The impact of acquired point mutations in the EGFR ectodomain on the binding ability of cetuximab.
a The location of acquired point mutations in EGFR. The structure of domain III (dark gray) in the EGFR ECD (white) was adapted from the Protein Data Bank (PDB accession code: 1YY9) and visualized by PyMOL. Single amino acid mutations that abrogate cetuximab or panitumumab binding are highlighted in cyan, except the S492R and G465R substitutions (purple). b The impact of acquired point mutations in the EGFR ECD on the binding ability of cetuximab and panitumumab (top panel). NIH3T3 cells stably expressing full-length EGFRWT or EGFRMut were treated with cetuximab (red) or panitumumab (blue) (1, 10 and 100 nmol/L) and further fluorescently labeled with a FITC-conjugated goat anti-human IgG (H + L) secondary antibody. Experiments were performed in triplicate. The bars indicate the mean ± SD values. S492R and G465R are the two most frequent mutations, according to the literature (bottom panel). c The reported crystal structure of the wild-type EGFR ECD/cetuximab Fab complex (PDB code: 1YY9) and the predicted structure of cetuximab in complex with EGFRS492R or EGFRG465R. Source data are provided as a Source Data file.