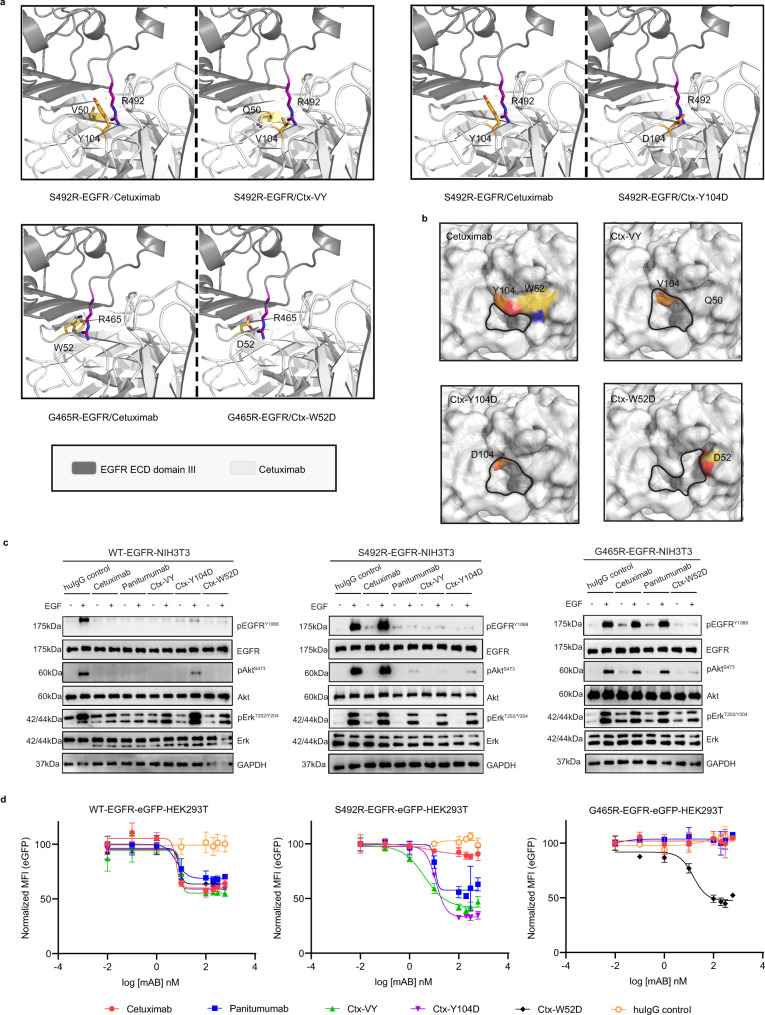
Fig. 5. The mechanisms of action by which the cetuximab variants in inhibit EGFRS492R or EGFRG465R activation.
a The modeled structural mechanisms by which the cetuximab variants restore binding with EGFRS492R or EGFRG465R. The critical residues in the cetuximab variants and the EGFR mutations are highlighted as colored sticks in the interface. The structures of VH domain in the cetuximab/cetuximab variants and EGFRS492R or EGFRG465R ECD domain III are shown in white and gray, respectively. b The central cavities (circled in black) in the paratopes of cetuximab (PDB code: 1YY9) and its variants. c The mechanism by which the cetuximab variants inhibit the ligand-induced activation of EGFRS492R or EGFRG465R, as shown by western blot analysis. Stable NIH3T3 cells were stimulated with EGF (0.5 nM) and immunoblotted for total and phosphorylated EGFR, Akt, and Erk (pEGFRY1068, pAktS473, and pErkT202/Y204). d The mechanism by which the cetuximab variants inhibit the ligand-induced receptor internalization and degradation of EGFRS492R or EGFRG465R, as shown by flow cytometry. The mean fluorescence intensity (MFI) of different stable HEK293T cells was determined after 48 h of preincubation with the indicated concentrations of antibodies. MFI values were normalized to the EGFR-eGFP signal in cells with no antibody treatment. All above assays were performed in triplicates. The bars indicate the mean ± SD values. Source data are provided as a Source Data file.