Figure 4 |. Cell line dependency and modular design of HDAC3-targeted degraders.
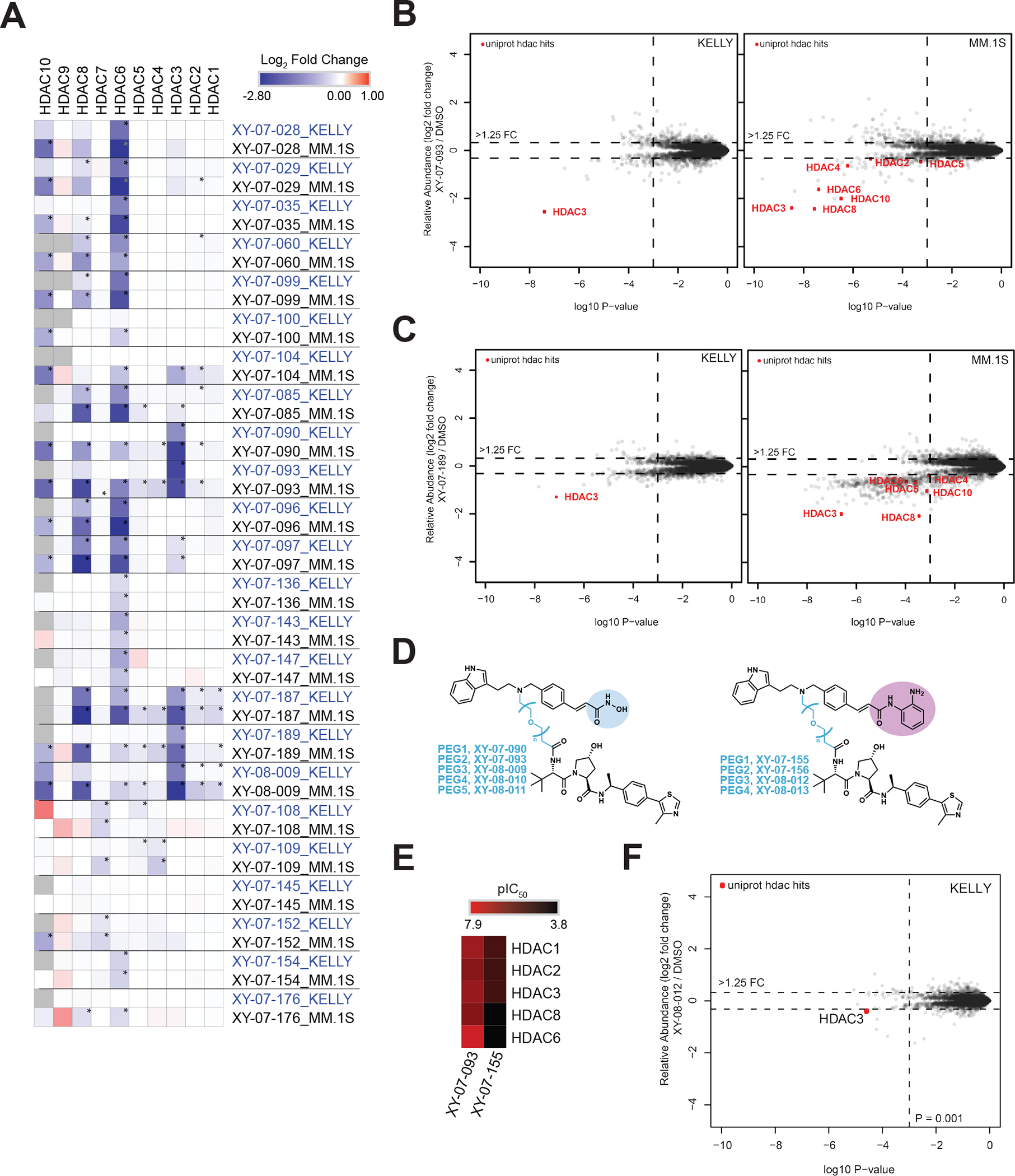
(A) Heatmap displaying the log2FC in relative abundance for HDACs in response to treatment with indicated degraders in KELLY and MM.1S cell lines. (B) Scatterplots depicting the log2FC in relative protein abundance in response to treatment with XY-07-093 in KELLY and MM.1S cells. (C) As in B but treated with XY-07-189. (D) Chemical structures of dacinostat-VHL1 degraders with hydroxamate or benzamide zinc-binding groups. (E) Heatmap displaying HDAC in vitro enzymatic inhibition data (pIC50) comparing HDAC selectivity for hydroxamate- (XY-07-093) and benzamide-based (XY-07-155) molecules. Inhibition data is from n = 1 ten-point titrations. (F) Scatterplot depicting the log2FC in relative protein abundance in response to treatment with XY-08–012. Proteomics data is from n = 1–2 biological replicates. * on proteomics heatmaps indicates a P-value < 0.001. See also Table S2–3, Figure S1, S3, S4.
