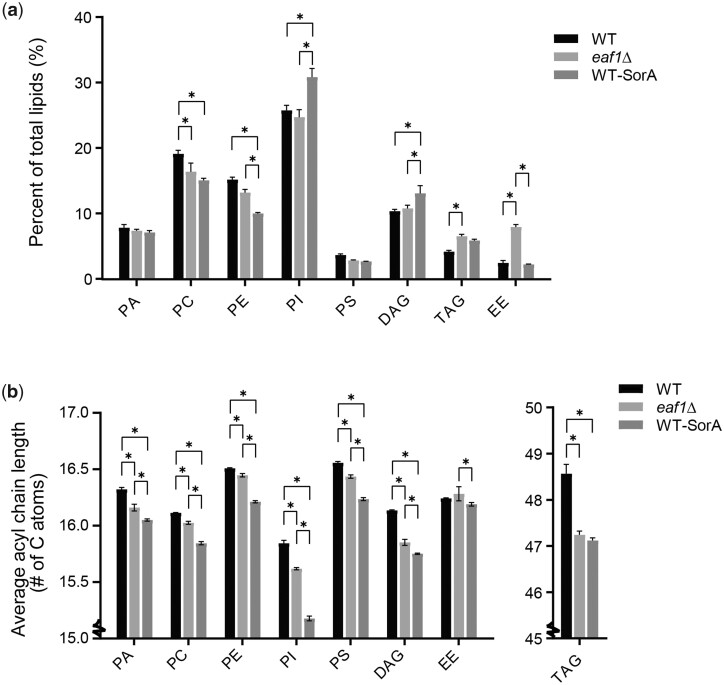
Fig. 7.
Lipidomic analysis of WT and NuA4 mutants. The lipid composition of WT, eaf1Δ, and Sor A treated WT yeast was assessed through lipidomics. WT, eaf1Δ, and Sor A treated cells were harvested in mid-log and a crude lysate was created through bead beating with water, lipids were then extracted using a chloroform/methanol procedure and Mass spec analysis was performed by Lipotype GmbH. In this figure are the most prevalent lipids identified: PA, Phosphatidylcholine (PC), Phosphatidylethanolamine (PE), Phosphatidylinositol (PI), Phosphatidylserine (PS), Diacylglycerol (DAG), Ergosteryl ester (EE), and Triacylglycerol (TAG). a) The composition of lipid classes broken down as a percent of the total lipids. A 2-way ANOVA analysis with a Tukey’s multiple comparisons test was performed and a * represents an adjusted P < 0.05. b) The average acyl chain length for each class of lipid was calculated from the lipidomic data. The Y-axis is broken to zoom in on the data and show the small but significant changes in chain length. A 2-way ANOVA analysis with a Tukey’s multiple comparisons test was performed and a * represents an adjusted P < 0.05.