Fig. 1.
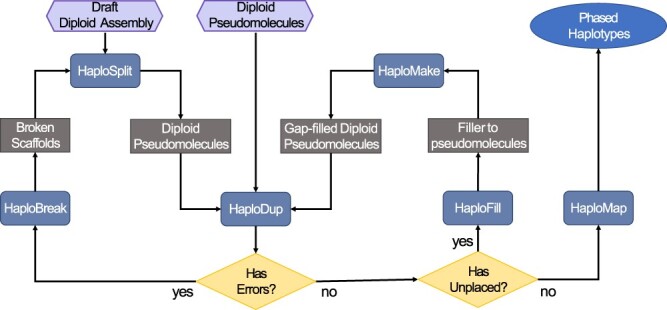
The HaploSync pipeline builds and refines haploid and diploid genome assemblies. The diploid-aware pipeline can deliver fully phased diploid pseudomolecules using a draft diploid assembly or diploid pseudomolecules as input. If draft sequences are used, Haplosplit first separates the haplotypes into 2 pseudomolecule sets. Pseudomolecules provided by the user or reconstructed with HaploSplit, then undergo quality control with HaploDup. If errors are found, input sequences can be edited with HaploBreak prior to rebuilding the pseudomolecules with HaploSplit. If no errors are detected and there are unplaced sequences, the pseudomolecule undergoes gap-filling with HaploFill. After each filling iteration, quality control can be performed with HaploDup. Finally, HaploMap can be used to identify colinear regions between pseudomolecules.
