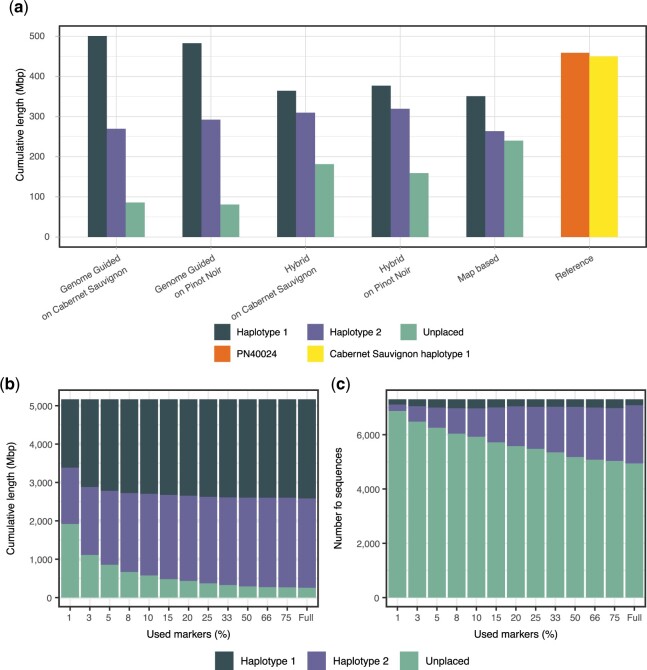
Fig. 4.
HaploSplit performance. a) The results of using different sources of external information and HaploSplit protocols for V. vinifera cv. Cabernet Franc cl. 04 (Vondras et al. 2021) assembly. Map-based assembly produces the largest first haplotype, but its overassembly occurs at the expense of the second haplotype’s completeness. A map-based approach is conservative and limited by the density of the markers. The hybrid approach recovers more sequences where the map is lacking information, without overassembling, and delivers a better reconstruction of both haplotypes. b) Effect of limited marker availability on overall assembly length tested on B. taurus Angus × Brahma (Koren et al. 2018; Low et al. 2020) by subsampling the genetic map. Longer sequences are more likely to contain a marker, making the first reconstructed haplotype most complete across all tests and with little variation in size. As the number of available markers increases and short sequences are included, the completeness of the second haplotype improves. c) Effect of limited marker availability on the number of placed sequences tested on B. taurus Angus × Brahma (Koren et al. 2018; Low et al. 2020) by subsampling the genetic map. Increasing the number of markers as fragmentation increases allows recruiting more sequences for scaffolding and improves completeness. Haplotype 1, with long sequences, shows little variation. In contrast, Haplotype 2 greatly benefits from increased marker density. The majority of sequences that remained unplaced are short and a small fraction of the genome’s length.