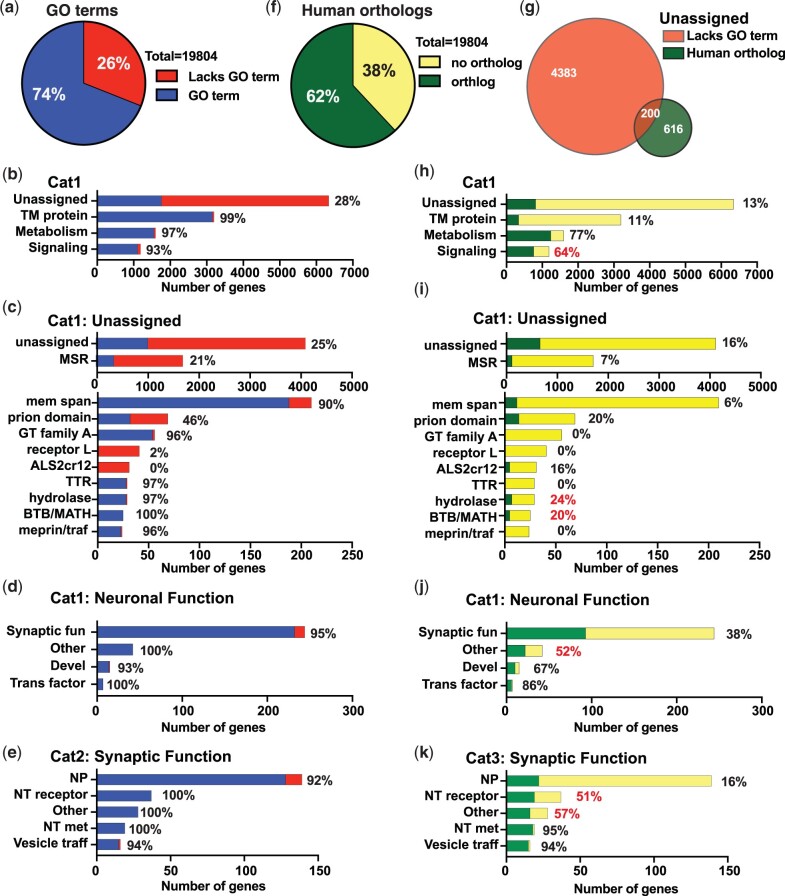
Fig. 2.
Unassigned genes in C. elegans include a subset with human orthologs. a) Pie chart of C. elegans protein-coding genes that are assigned GO terms in the ParaSite Biomart. b) Breakdown of WormCat Cat1 level categories with numbers of genes annotated by GO. Cat2 GO breakdown of Unassigned (c) and Neuronal function (d) with Cat3 level categories of Synaptic Function. Additional categories are in Supplementary Fig. 2, a–d. f) Pie chart of C. elegans protein-coding genes designated as having human orthologs in the ParaSite Biomart. g) Venn diagram showing the overlap between Unassigned genes lacking GO terms and those with human orthologs. h) Breakdown of WormCat Cat1 level categories with numbers of genes with human orthologs. Cat2 human ortholog breakdown of Unassigned (i) and Neuronal function (j) with Cat3 level categories of Synaptic Function (k) (Supplementary Fig. 2, e–h). Red percentages denote categories with substantially more human orthologs in a genome-wide BLASTP comparison of each C. elegans gene with the human genome (see Supplementary Table 4). TM, transmembrane; msr, multiple stress-regulated; mem span, membrane-spanning; GT family A, glucosyltransferase family A; TTR, TransThyretin-Related family domain; BTB/MATH, BR-C, ttk, and bab/meprin and TRAF homology; Synaptic fun, Synaptic function; Devel, Development; Trans, Transcription; NP, Neuropeptide, NT receptor, Neurotransmitter receptor; NT met, Neurotransmitter metabolism; Vesicle traff, Vesicle traffic.