Fig. 5.
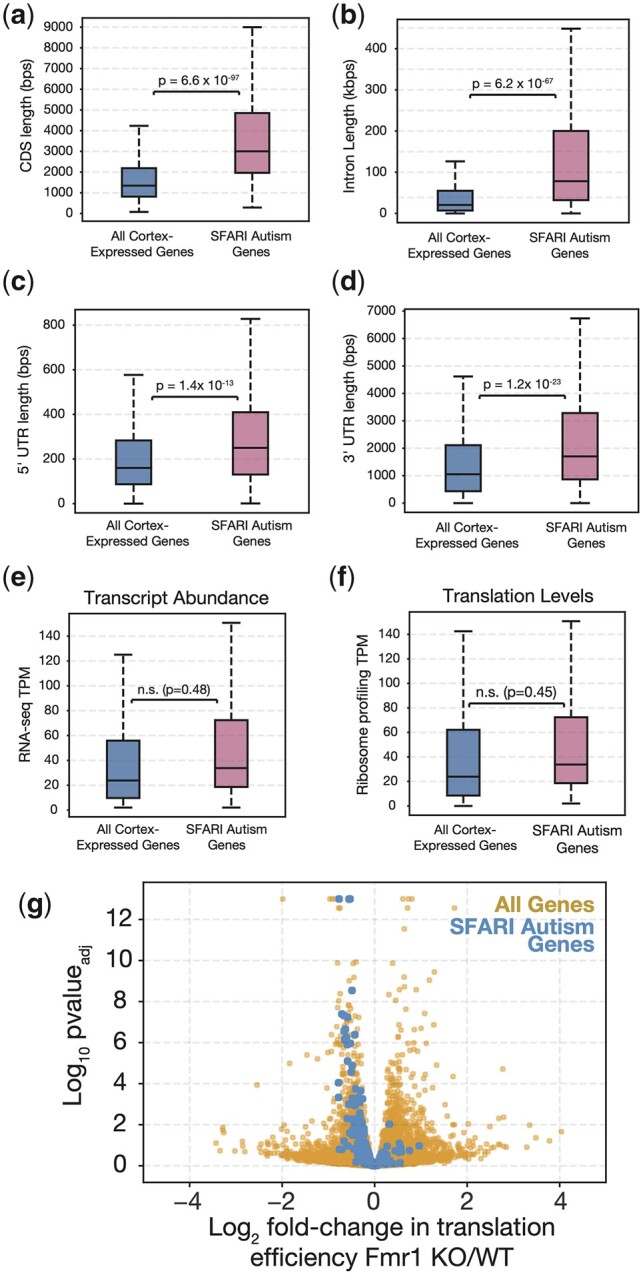
SFARI autism genes have longer open-reading frames, UTRs, and introns than average. Box plots of length distributions showing increased a) coding sequences lengths, b) 5′ UTR lengths, c) intron lengths (summed across all introns), and d) 3′ UTR lengths specifically for SFARI Class I and Class II ASD genes (magenta) as compared to all genes expressed in the juvenile mouse cortex (blue). No significant differences were detected between the distributions of transcript levels (e) or translation levels (f) specifically for ASD-relevant genes vs all cortex-expressed genes as detected by mRNA sequencing and ribosome footprinting, respectively. g) Volcano plot showing a concordant downregulation in the translation efficiency of SFARI Class I and Class II autism genes as compared to all neuronally expressed genes.
