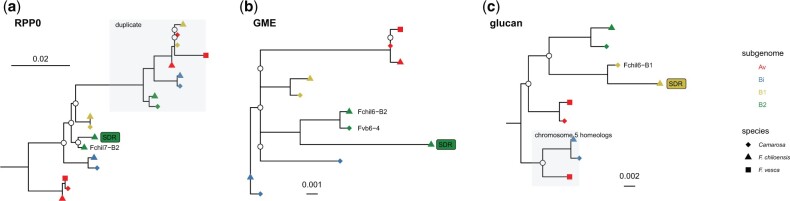
Fig. 6.
Maximum likelihood phylogenetic trees of (a) ribosomal protein P0 (RPP0), (b) GDP-mannose-3′,5′-epimerase (GME), and (c) glucan endo-1,3 beta-glucosidase (glucan). Nodes with bootstrap support <80 are represented by white circles. The copy of the genes present in the female-specific SDR is labeled “SDR” with a rectangle filled with the subgenome color of its original autosomal copy (Av = red, B1 = yellow, B2 = green, Bi = blue). Gray areas depict genes belonging to the gene families but not homeologous to the original autosomal copy of the SDR genes. The chromosomal location is given for the sister copies of the SDR genes. Tree visualization was done in R version 4.1.0 (R Core Team 2021) using ggtree v.3.0.4 (Yu 2020). The complete phylogenies are given in the Supplementary Figs. 13–15.