Figure 1. A proteome-wide map of pathogenic mutations in condensate-promoting features. See also Figures S1–S2 and Table S1.
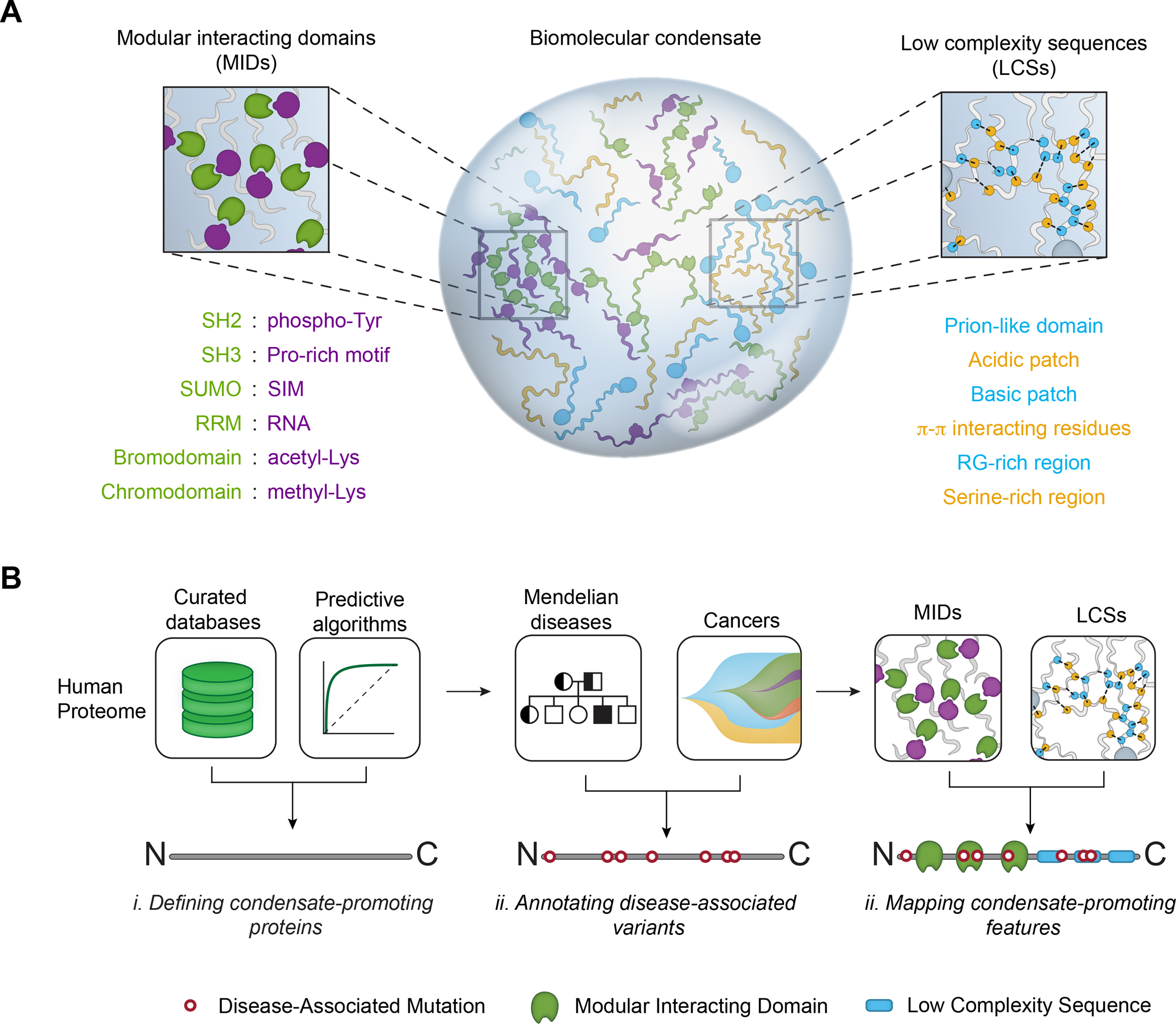
A. Multivalent interacting features in proteins that promote biomolecular condensate formation, including modular interacting domains (MIDs, left, green and purple) and low complexity sequences (LCSs, right, blue and green).
B. Approach to generate a map of pathogenic mutations that affect condensate-promoting features across the proteome (see also Figure S1A). MIDs and LCSs were mapped across the proteome (left, top) and used to define multivalent proteins (middle). Mendelian and cancer variants were mapped across the proteome (left, bottom), in particular on across the set of multivalent proteins (middle), to identify pathogenic mutations that affect MIDs and LCSs (Methods). The approach allows analysis of diseases, condensates, and mutational signatures associated with pathogenic mutations that affect condensate-promoting features in multivalent proteins (right).
