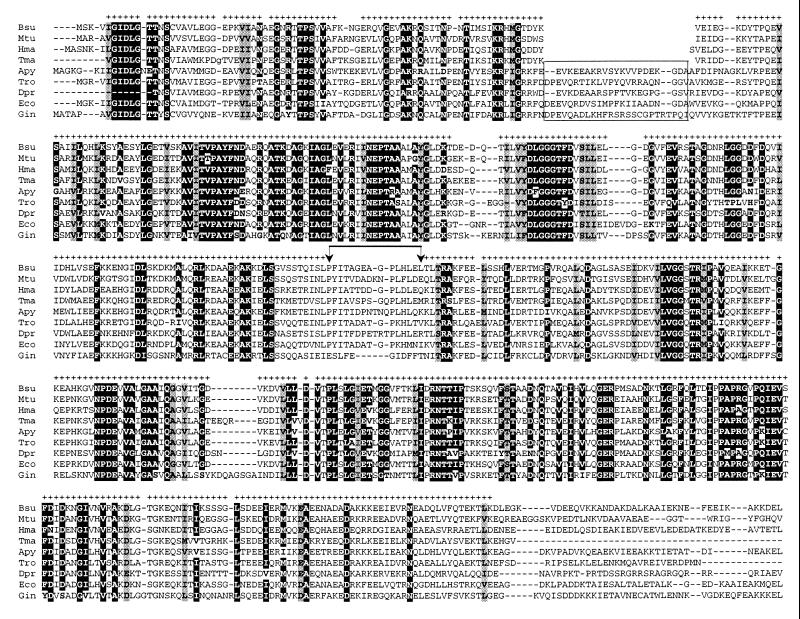
FIG. 1.
Abridged alignment of the A. pyrophilus and T. maritima HSP70 sequences with homologues from the low-G+C gram-positive bacteria (Bsu), the high-G+C gram-positive bacteria (Mtu), the archaea (Hma), the green nonsulfur bacteria (Tro), the deinococci (Dpr), the proteobacteria (Eco), and the eucarya (Gin). Plus signs indicate amino acid positions selected for construction of the phylogenetic trees shown in Fig.s 3 to 5; highlighted blocks indicate positions occupied by an identical amino acid in more than 90% of the 70 aligned sequences; shaded areas indicate positions occupied by similar amino acids (ILVM, DEKRH, ST, GA, FYW, NQ) in 100% of the seventy sequences. For reasons of space, the C-terminal portion of the alignment (comprising residues not selected for tree reconstruction) is not shown. Species abbreviations are listed in the legend to Fig. 2. Vertical arrows delimit a sequence region that could not be confidently aligned between the procaryotic and eucaryotic HSP70s. The boxed region in the top block delimits the insertion segment found in many HSP70s.