Abstract
Background
Pro-inflammatory cytokines are dysregulated in Crohn’s disease (CD) and could serve as surrogate markers to improve diagnostic and therapeutic approaches, potentially addressing an unmet need. We profiled circulating biomarkers and whole blood transcriptional pathway activity to identify those associated with CD using data from the phase 2 FITZROY study with filgotinib, an oral preferential janus kinase-1 inhibitor.
Methods
Patients with serum and whole blood samples taken from the induction period were included. Serum cytokines were measured (ELISA), whole blood RNA sequenced, and stool samples taken to measure fecal calprotectin (FC). Spearman’s Rank correlations were assessed between biomarkers and baseline disease activity; post-treatment endoscopic improvement was measured by the Simplified Endoscopy Score for CD (SES-CD), FC and the Crohn’s Disease Activity Index. Effect of filgotinib on circulating biomarkers was also evaluated.
Results
Serum biomarkers (n = 168) and whole blood RNA sequencing (n = 104) were assessed. Moderate correlation between serum analytes with SES-CD and FC was noted; most highly correlated were acute phase proteins CRP (rho = 0.35 [SES-CD] and 0.47 [FC]), serum amyloid A (rho = 0.40 and 0.39, respectively) and pro-inflammatory cytokines interleukin (IL)-6 (rho = 0.31 and 0.30, respectively), IL-22 (rho = 0.36 and 0.35, respectively), and oncostatin M (rho = 0.35 and 0.33, respectively). Filgotinib treatment was associated with reduction of many candidate biomarkers, particularly in patients with treatment response. Early changes in IL-6 and IL-10 may be prognostic for endoscopic response.
Conclusions
Several circulating factors with potential as CD activity biomarkers were identified. Larger studies are necessary to investigate the best utility of these markers for CD.
Keywords: IBD, filgotinib, Crohn’s disease, JAK1, blood
Introduction
Crohn’s disease (CD) is a debilitating, incurable, immune-mediated, chronic, idiopathic inflammatory bowel disease (IBD) that can affect the entire gastrointestinal tract.1,2 Symptoms may include abdominal pain, diarrhea, fatigue, weight loss, fever, and chills, and the condition can sometimes lead to life-threatening complications.1,3 Around half of all patients with CD require surgery within 10 years of diagnosis and require chronic usage of corticosteroids and other immunosuppressive medications.2
Treatment of CD is based on the severity of the symptoms and underlying disease phenotype.1 However, options for assessing disease activity and severity are limited and often invasive. Traditionally, they involve macroscopic examination of the appearance of the intestinal mucosa via video endoscopy and histopathological examination of the resulting tissue biopsy samples collected; these methods carry risk to the patient and are expensive and time-consuming.4–6 Alternatives are therefore needed. The identification of cytokines or genes involved in disease pathology that could be used as robust surrogate biomarkers of disease activity, especially circulating biomarkers that may correlate to activity at the tissue level, would be of benefit to the diagnostic and therapeutic approaches to CD.5–7
The study of cytokines associated with the immunoregulatory pathways of CD has helped to develop new modalities for diagnosis, prognosis, and treatment.7 Changes in CD management emerged with the introduction of biologic therapies targeting pro-inflammatory cytokines that are believed to be pivotal in the pathophysiology of CD.8,9 These therapies have provided substantial benefit in outcomes for patients with CD, improving symptoms, quality of life and resolution of inflammation.10,11 Biologic therapy with tumor necrosis factor (TNF) blockers has been the gold standard treatment for moderate to severe CD for many years.9,11
Many studies have been conducted to identify specific biomarkers that reflect the level of mucosal inflammation in patients with CD; blood C-reactive protein (CRP), a general, nonspecific marker of systemic inflammation, and fecal calprotectin (FC), which more specifically reflects neutrophil migration and activation in the intestinal mucosa, are commonly used as surrogate markers of disease activity in CD.4–6 The CALM study showed that better clinical and endoscopic outcomes could be achieved using CRP and FC in combination with clinical symptoms to decide on the timing of treatment escalation.12 However, both CRP and FC show inconsistent correlation with mucosal inflammation when compared with endoscopic assessments of disease activity.6 These shortcomings mean that there is still a tangible need for novel biomarkers that reflect disease activity in CD, particularly in patients with subclinical disease, and for biomarkers that enable a noninvasive assessment of response to treatment.6,7
A large number of genes (~300) have been linked to the risk of CD, many of which can be associated with changes in immune pathways, so a broad panel of serologic and immunologic biomarkers other than FC and CRP could be used to investigate the disease activity by comparison with clinical disease metrics.7,13 Although increased production of cytokines and mediators at the site of inflammation likely facilitate pathological effects in CD,7,14 their systemic increases may provide a surrogate measure of intestinal inflammation.
In terms of treatment of CD, not all patients respond well to anti-TNF-α and a search for novel treatments with a different mode of action is ongoing.15 Janus kinases (JAKs) are tyrosine kinases located in the cytoplasm that transduce signals following the binding of cytokines to their cognate membrane receptors.15 Janus kinases phosphorylate the signal transducers and activators of transcription (STATs), which then dimerize and translocate to the nucleus where they activate gene transcription.16,17 Janus kinase-1 is essential for signaling for certain type 1 and type 2 cytokines, and type 1 and 2 interferons (IFNs), many of which have been reported to be elevated in patients with CD.14,18,19 The JAK inhibitors represent a relatively new and promising class of drugs being investigated for the treatment of patients with CD and other chronic inflammatory disorders.16
Filgotinib is an oral selective JAK1 inhibitor.16,20 The efficacy of filgotinib to treat moderately to severely active CD was investigated in the phase 2, multicenter, randomized, double-blind, placebo-controlled FITZROY study (NCT02048618). It was found that significantly more filgotinib-treated patients with CD achieved clinical remission (Crohn’s Disease Activity Index [CDAI] <150) compared with placebo-treated patients (47% vs 23%, P = .0077).15 Filgotinib is expected to reduce cytokine signaling via inhibition of the JAK1-STAT pathway in the inflamed mucosa and thereby dampen inflammation, leading to endoscopic improvement in CD.15
The objective of the present study was to identify circulating biomarkers and transcriptional pathway activities in whole blood samples drawn from the FITZROY study that could be used as surrogate markers of disease activity assessed at the site of inflammation. In addition, the study would assess the response to treatment with filgotinib and the effect of prior anti-TNF-α therapy.
Materials and Methods
Study Design and Patient Disposition
Evaluation of disease-related biomarkers and the effect of filgotinib was a planned analysis of the FITZROY study. Full details of the study design, patients, inclusion/exclusion criteria, and the results from FITZROY have been described in detail elsewhere.15 Briefly, the study included 2 parts, each spanning a 10-week duration. In part 1 (induction), patients were randomized (3:1) to receive 200mg of filgotinib once a day or placebo. In part 2, following the 10-week induction period, patients were rerandomized to receive either 200mg of filgotinib once a day, 100mg of filgotinib once a day, or placebo for an additional 10 weeks. Serum and whole blood samples collected from induction baseline through to week 10 (part 1) of FITZROY were used in the current analysis.
Ethical Considerations
The study protocol was reviewed and approved by the relevant independent ethics committees for each center and was in accordance with the Declaration of Helsinki,21 Good Clinical Practice guidelines,22 and all applicable regulatory requirements. All patients provided written informed consent.
Biomarker Sampling
Serum samples were collected at baseline and at weeks 2, 4, and 10 following treatment initiation. Whole blood samples were collected into PAXgene tubes at baseline and week 10.
A stool sample was collected at every visit (except follow-up) to measure the levels of FC. The analysis of FC was made using an enzyme-linked immunosorbent assay (ELISA) performed by the central laboratory (BARC, Belgium).
Serum Analyte Detection
Serum cytokines were measured by ELISA at several laboratories. Analytes for which more than 10% of samples had cytokine concentrations below the lower limits of quantitation (LLOQ), which is the lowest amount of an analyte in a sample that can be quantitatively determined with suitable precision and accuracy, were excluded from statistical analyses but are listed in Supplementary Table 1. The following analytes were included in the biomarker analysis: interleukin (IL)-6, IL-8, IL-10, IL-17A, vascular endothelial growth factor A (VEGFA), interferon gamma (IFN-γ), serum amyloid A (SAA), glycoprotein (GP) 130 and serum calprotectin (CALP), which were all measured by Pacific Biomarkers; eotaxin, transforming growth factor beta 1 (TGFb1) and IL-22, measured by ABL (Lyon, France); degraded type 3 collagen (C3M), propeptide of type 3 collagen (PRO-C3) and degraded type 4 collagen (C4M), measured by Nordic Bioscience (Herlev, Denmark); and oncostatin M (OSM), which was measured by Gilead Sciences, Foster City, CA.
The central laboratory measured albumin levels using colorimetry/bromocresol green (BCG) dye binding, CRP using immunoturbidimetry, and hemoglobin using spectrophotometry (Supplementary Table 1).
Whole Blood RNA-Seq Library Preparation and Gene Expression Quantitation
RNA was isolated from whole blood using a PAXgene 96 Blood RNA Kit (Qiagen, Germantown, MD, USA), and globin mRNA was removed with GlobinClear Human Kit (ThermoFisher, Waltham, MA, USA). RNA-seq libraries were prepared with TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA, USA) and were sequenced on Illumina HiSeq 2500. Samples from 3 patients were removed due to quality control failure. Gene expression levels were quantitated from RNA-seq raw sequence files (FASTQ) using Salmon.23
Pathway Activation in Whole Blood
Hallmark pathway24 activity scores were calculated from whole blood RNA-seq using single sample Gene Set Enrichment Analysis (ssGSEA).25 Pathway gene sets were downloaded from MSigDB.26 For each pathway, activity scores were standardized by the mean and standard deviation of the activity scores of the baseline samples. For each Hallmark pathway, the proportion of genes in the pathway gene set that was expressed in whole blood RNA-seq was calculated; a gene was considered expressed if at least 10% of samples had counts per million (cpm) >1. Pathways for which less than 60% of the component genes were expressed were removed from the analysis.
Assessment Methods and Statistical Analyses
Baseline correlation analysis
Analytes and pathway activity scores were correlated with clinical disease activity metrics in order to identify peripheral markers associated with disease severity at baseline. The clinical metrics included SES-CD, a validated index of endoscopic disease activity that allows for objective assessment of response to therapy4; FC, which is a marker of neutrophil activation used clinically for the diagnosis and ongoing assessment of IBD4; and the CDAI score, a composite score commonly used to assess CD activity.27 Two subcomponents of the CDAI, the Stool Frequency Subscore (SFS) and Abdominal Pain Subscore (APS), which are measures of CD symptoms, were also used.27
Comparisons included baseline correlation between serum biomarkers and the clinical metrics, baseline correlation between whole blood pathway activities and clinical metrics, and between gene expression. All correlations were calculated using Spearman’s Rank method, then displayed in heatmaps to illustrate the strength of the correlations. A cutoff of 0.3 was applied for the absolute value of the correlation coefficient as a minimum strength of the correlation.
Treatment effect analysis
The effect of filgotinib and placebo treatment on circulating biomarkers was assessed by examining the change in concentration of serum analytes from baseline to week 2, week 4, and week 10 for endoscopic responders (who were defined as a reduction from baseline of SES-CD score ≥50%) and nonresponders. To assess the log-fold changes in protein biomarker concentrations from baseline during treatment, a mixed effects model (R packages emmeans and lme4) was used with terms for treatment, visit, and the interaction between the treatment and the visit, with baseline biomarker level modeled as fixed effects covariate and subject identification as a random effect. Corrections for multiple testing were performed using the Benjamini-Hochberg method.28 A false-discovery rate (FDR) or P value cutoff of 0.05 was used to define statistical significance.
The effect of filgotinib treatment on whole blood gene expression and pathway activities were also investigated, examining changes in gene expression levels or pathway activity scores between baseline and week 10. Differential gene expression analysis between week 10 and baseline was performed using limma/voom R package.29,30 A design matrix for the linear model was constructed as an interaction between the visit and treatment. Differential pathway activity analysis between week 10 and baseline was performed using limma R package29 using the same design matrix used for differential gene expression. Corrections for multiple testing were performed using the Benjamini-Hochberg method.27 A false-discovery rate (FDR) or P value cutoff of 0.05 was used to define statistical significance.
Early change in biomarkers for endoscopic response
Logistic regression was performed to identify biomarkers whose early change (week 2) from baseline may be prognostic or predictive of endoscopic response at week 10, using terms for treatment, sex, age, baseline SES-CD score, and baseline biomarker level as covariates. For each biomarker, patients were divided based on whether the biomarker level increased or decreased. Due to the small sample size for the placebo arm, the logistic regression analysis for biomarkers predictive of filgotinib response was not sufficiently powered. A model with an interaction term between change in biomarker level and treatment was tested, but as expected, there were no significant biomarkers. Thus, a model without the interaction term was used. Additionally, logistic regression was used to determine whether changes in levels of combinations of biomarkers impacted endoscopic response at week 10, using terms for biomarker change (increase/decrease), treatment, sex, age, and baseline SES-CD score.
Impact of prior anti-TNF-α therapy on treatment effects
An analysis was performed to understand if there were differences between peripheral markers associated with disease activity at baseline, or those that reflect endoscopic improvement with filgotinib treatment in patients with or without a prior history of anti-TNF-α treatment. Baseline correlation with SES-CD was calculated using the Spearman’s Rank correlation method. Statistical significance of change from baseline in biomarker levels was tested using a mixed effects model for treatment effect analysis with subgroups naïve or experienced to anti-TNF-α; this was corrected for multiple testing using the Benjamini-Hochberg method.
Results
Patient Disposition
In the induction phase (part 1) of the FITZROY trial, from which the current samples were obtained, patients were enrolled and randomly assigned to receive 200mg of filgotinib once a day (n = 130) or placebo (n = 44).15 Biomarker and whole blood RNA-seq data were generated from all patients who provided samples; biomarker sample collection was optional, and as a result, not all patients provided samples. Serum biomarkers were measured, and stool samples taken from 168 patients, whereas whole blood RNA-seq was generated from 104 patients, minus the 3 quality control failures (n = 101). The demographics, baseline characteristics, and clinical response of the serum biomarker and whole blood analysis sets were not statistically different from the full randomized set (Supplementary Table 2).
Baseline Correlation Analyses
Baseline correlation between serum analytes and clinical metrics
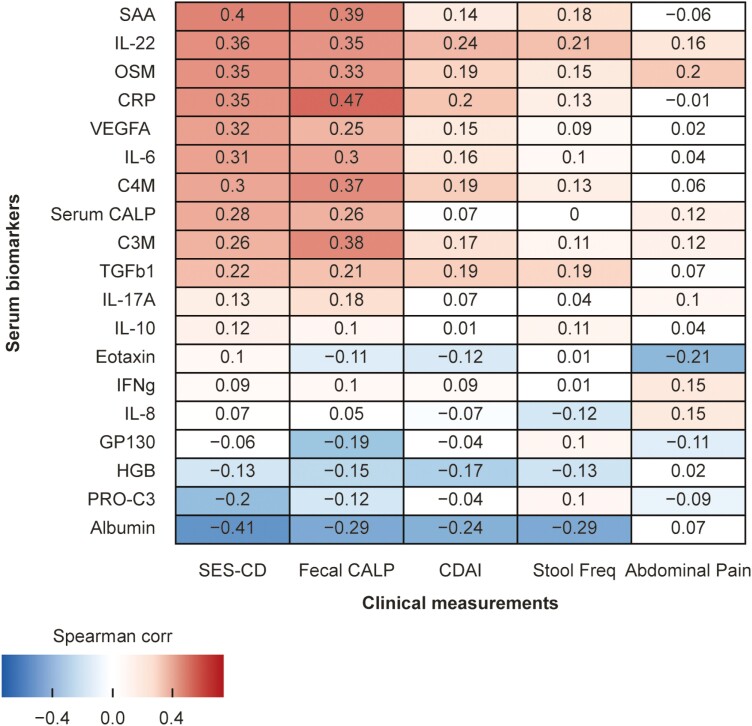
For all analytes used in the analyses, less than 5% of samples were below the LLOQ. Among the 5 metrics, CDAI, SFS, and APS had a weak or no correlation (|rho| < 0.3) with all serum analytes, whereas SES-CD and FC showed moderate correlation (0.3< |rho| <0.5) with several serum analytes at baseline (Figure 1 and Supplementary Figure 1A).
Figure 1.
Correlation between serum biomarker levels and clinical measurements at baseline. Red indicates a positive correlation and blue indicates negative correlation.
The following are the serum analytes that were moderately positively correlated with SES-CD: the acute phase proteins CRP (rho = 0.35) and SAA (rho = 0.40); the pro-inflammatory cytokines IL-6 (rho = 0.31), OSM (rho = 0.35), and IL-22 (rho = 0.36); and an angiogenesis marker, VEGF-A (rho = 0.32). Albumin (rho = -0.41) was moderately negatively correlated with SES-CD. Serum analytes positively correlated with FC included CRP (rho = 0.47), SAA (rho = 0.39), IL-6 (rho = 0.30), OSM (rho = 0.33), IL-22 (rho = 0.35), and the matrix metalloproteinase (MMP)-mediated type 3 and type 4 collagen degradation markers C3M (rho = 0.38) and C4M (rho = 0.37). Inflammatory cytokines such as IFN-γ, IL-17A, and IL-10 were not correlated with clinical metrics at baseline.
Baseline correlation between clinical metrics and whole blood gene expression or pathway activities
The Simplified Endoscopy Score for CD was positively correlated (rho ≥ 0.3) with whole blood mRNA expression of 212 genes, which included immune-related genes such as suppressor of cytokine signaling 3 (SOCS3; rho = 0.56) and OSM (rho = 0.50). The SES-CD was negatively correlated (rho ≤ −0.3) with expression of 1283 genes; these included many genes related to RNA transcription and proliferation. Similarly, there were 111, 59, and 10 genes with expression positively correlated with FC, SFS, and CDAI, respectively; there were 249, 160, 12, and 9 genes whose expressions were negatively correlated with FC, SFS, CDAI, and APS, respectively.
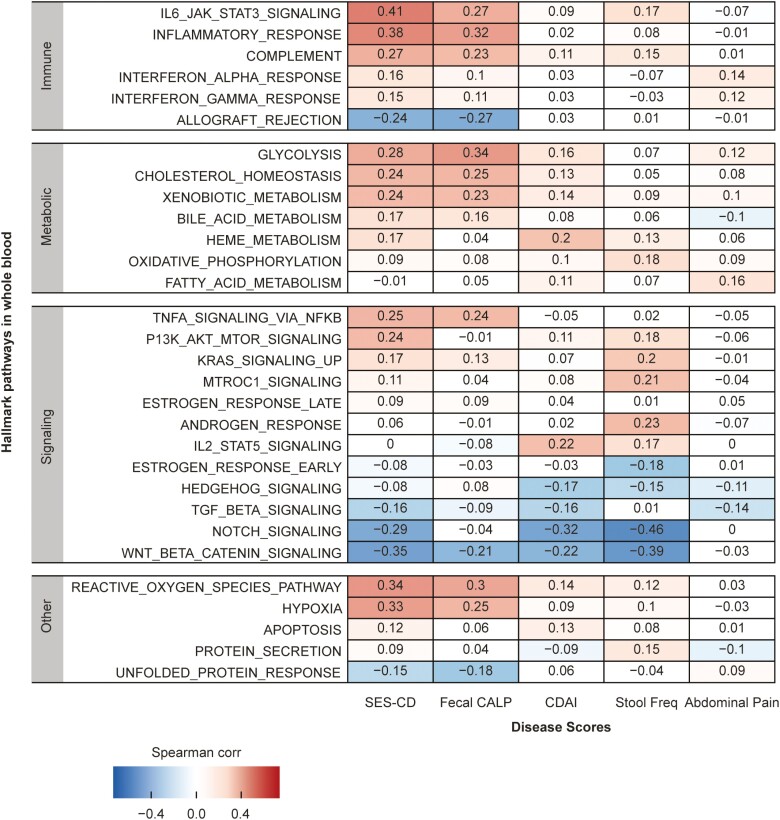
Gene expression mapping into the canonical Hallmark pathways showed that among the 5 metrics, SES-CD and FC showed moderate correlation with several pathway activity scores but not CDAI and APS (Figure 2 and Supplementary Figure 1B). Pathways that were moderately positively correlated with SES-CD included the immune-related pathways “IL6/JAK/STAT3 signaling” (rho = 0.41), “inflammatory response” (rho = 0.38), “reactive oxygen species” (rho = 0.34), and “hypoxia” (rho = 0.33). The “wingless-related integration site (WNT) beta catenin signaling” pathway was moderately negatively correlated with SES-CD (rho = -0.35). Pathways that were moderately correlated with FC included “inflammatory response” (rho = 0.32) and “reactive oxygen species” (rho = 0.30). Pathways with activity scores that positively correlated to SES-CD and FC were also moderately correlated with several serum biomarkers including acute phase proteins and systemic inflammation markers (Supplementary Figure 2).
Figure 2.
Correlation between Hallmark pathway activity scores derived from mRNA expression and clinical metrics at baseline. Heatmap shows pathways in Immune, Metabolic, Signaling, and Pathway categories only. Red indicates positive correlation and blue indicates negative correlation. Abbreviations: CALP, calprotectin; CDAI, Crohn’s disease activity index; SES-CD, Simple Endoscopic Score for Crohn’s Disease.
Treatment Effect Analyses
Filgotinib treatment effect on serum analytes
Consistent with the endoscopic response criterion (SEC-CD decrease at least by 50%), in the filgotinib-treated group, SES-CD at week 10 was significantly lower in the responder group than the nonresponder group (median [Q1, Q3]: 5 [3, 7.5] vs 12 [8, 18], respectively; P = 5.20e-12). Although not statistically significant, the decrease in FC from baseline to week 10 among patients using filgotinib was also numerically greater in patients with endoscopic response (−176 [−561, 59]) compared with the nonresponders (−75 [−202, 69]).
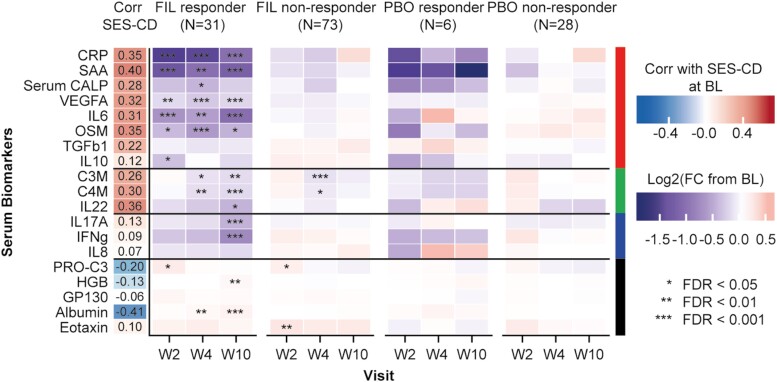
For the endoscopic responders, a statistically significant reduction of many serum analytes across visits was observed in the filgotinib treatment arm (Figure 3 and Supplementary Figure 1C). In the placebo treatment arm, for the endoscopic responders there were no analytes with statistically significant changes, though this may have been due to a combination of the small sample size (n = 34) and small effect size in the placebo arm. However despite these limitations, the trend in direction of change for the placebo arm did seem to be similar to the filgotinib arm. There were very few analytes with statistically significant changes among the filgotinib nonresponders, and none were significant among the placebo nonresponders.
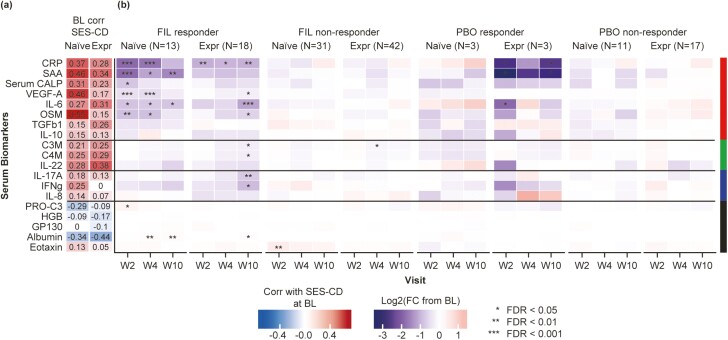
Figure 3.
Correlation of serum analytes with SES-CD across the study period in both the filgotinib and PBO arms (mixed effects model). Log2 (fold change from BL) was determined by comparing levels at the post-treatment visit to BL and displayed as a heatmap for endoscopic responders and nonresponders in FIL and PBO treatment arms. Red indicates increase from BL and blue decrease from BL. Statistical significance of fold change from BL was determined using mixed effects model, and corrected for multiple testing using Benjamini-Hochberg method. Abbreviations: BL, baseline; Corr, correlation; FC, fold-change; FDR, false-discovery rate; FIL, filgotinib; PBO, placebo; SES-CD, Simple Endoscopic Score for Crohn’s Disease; W, week.
All serum analytes with a positive correlation (rho > 0.3) to SES-CD at baseline were reduced by filgotinib treatment, but there were notable differences in the kinetics of changes among analytes, as well as between endoscopic responders and nonresponders. Filgotinib treatment was associated with an early (week 2) reduction of acute phase proteins (CRP, SAA) and pro-inflammatory cytokines (IL-6, OSM) of greater magnitude in endoscopic responders than in nonresponders (Figure 3). The early reduction in CRP, SAA, VEGFA, IL-6, and OSM was sustained at week 4 and week 10. In contrast, IL-10, which was significantly reduced at week 2 in endoscopic responders, returned to near baseline levels at week 4 and week 10. Degraded type 3 collagen, C4M, and IL-22, which were positively correlated with the intestinal inflammation marker (FC) at baseline, showed a delayed (week 4) reduction by filgotinib treatment. Interferon-γ and IL-17, which both showed a weak correlation with SES-CD or FC at baseline, were reduced by filgotinib treatment in a progressive decline from baseline levels over time (week 10). This treatment effect was observed only in endoscopic responders.
Filgotinib treatment effect on whole blood pathways
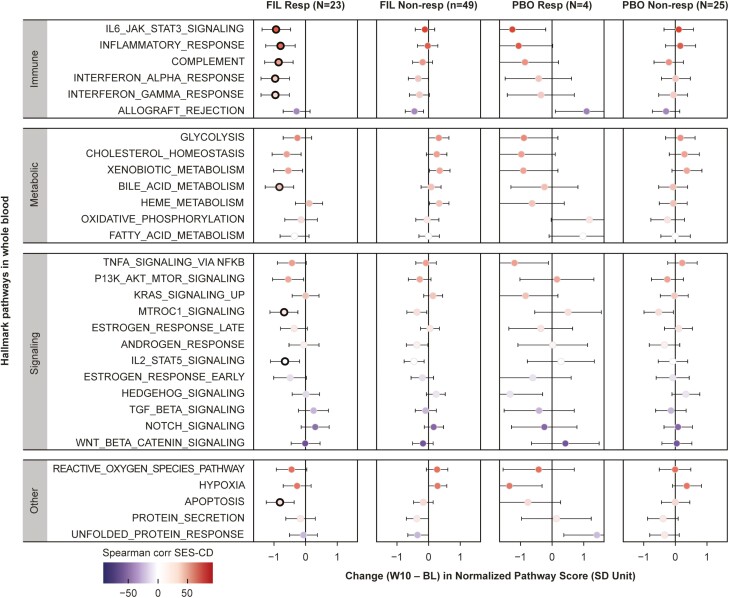
Changes in pathway activity scores between baseline and week 10 are shown in Figure 4. Among the pathways that were correlated with baseline SES-CD (Figure 2), systemic inflammation pathways (“IL6/JAK/STAT3 signaling,” “inflammatory response”) were significantly reduced with filgotinib treatment in endoscopic responders only, a pattern similar to pro-inflammatory cytokines IL-6 and OSM. Although not correlated with endoscopic disease severity at baseline, the IFN pathways (“IFN-α response” and “IFN-γ response”) showed significant decrease in activity with filgotinib treatment, consistent with reduction of IFN-γ in serum. Other pathways such as “IL2/STAT5 signaling,” “mTORC1 signaling,” “bile acid metabolism,” and “apoptosis” were also significantly reduced with filgotinib treatment to a greater degree in endoscopic responders vs nonresponders. There were no statistically significant changes from baseline in the placebo treatment arm in either the responder or nonresponder groups.
Figure 4.
Correlation of change from baseline to week 10 in Hallmark pathway scores with SES-CD for both the filgotinib and PBO arms. The pathway score is standardized using the mean and standard deviation of ssGSEA scores of baseline samples. Normalized score of 1 indicates a standard deviation of baseline distribution increase from baseline, and similarly, −1 indicates a standard deviation decrease. Each dot is colored by the Spearman’s Rank correlation coefficient with SES-CD at baseline; black encircled dots denote statistically significant (FDR < .05) pathway change from baseline. Abbreviations: BL, baseline; Corr, correlation; FIL, filgotinib; PBO, placebo; SES-CD, Simple Endoscopic Score for Crohn’s Disease; ssGSEA, single sample Gene Set Enrichment Analysis; W, week.
Early Change in Biomarkers for Endoscopic Response
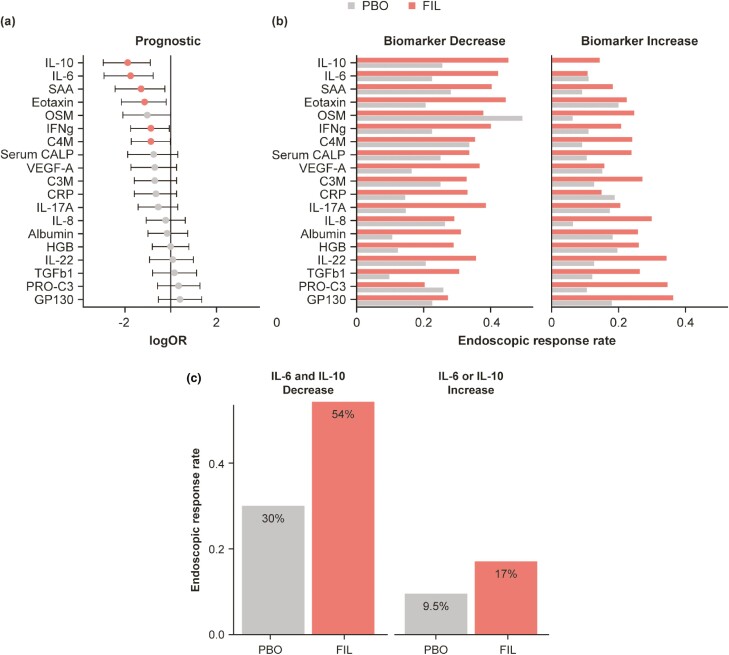
Decrease at week 2 in IL-6 (odds ratio [OR], 0.17; P = .0012) or IL-10 (OR, 0.16; P = .00034) level was prognostic of endoscopic response (Figure 5A and B). For patients with a decrease in IL-10, response rates at week 10 were 40.7% for those receiving filgotinib and 22.7% for those receiving placebo. In contrast, endoscopic response rates in patients who experienced an increase in IL-10 level were 12.5% and 0%, respectively. Similarly, response rates in patients with early decrease in IL-6 level were 37.9% and 20% for filgotinib and placebo arms, respectively; in patients with increased IL-6 levels, the rates were 9.3% and 9.5%, respectively. Despite these differences, the change at week 2 in IL-10 or IL-6 were not predictive of filgotinib response using the full interaction model. Biomarkers that were found to be marginally significantly prognostic were SAA (OR, 0.27; P = .019), C4M (OR, 0.42; P = .048), IFN-γ (OR, 0.42; P = .044), and Eotaxin (OR, 0.31; P = .019). The concordance of the change (increase/decrease) in IL-6 and IL-10 levels was assessed and found to be poor, with Cohen kappa statistic of 0.25 (percent concordance 63%). The combined early decreases in IL-6 and IL-10 were prognostic of endoscopic response (OR, 0.15; P = .000094), compared with IL-10 (OR, 0.16) or IL-6 (0.17) individually (Figure 5C).
Figure 5.
Early change in biomarkers for endoscopic response. Logistic regression of early change (W2) in serum biomarkers levels on endoscopic response at W10. A, Dot is the average log (odds ratio) and the lines are 95% confidence intervals. Red indicates P < .05. B, Endoscopic response rate for biomarker increase or decrease subgroups for FIL or PBO. C, Endoscopic response rates for a subgroup with decreases in both IL-6 and IL-10 or increase in IL-6 or IL-10 when treated with FIL or PBO. Abbreviations: FIL, filgotinib; PBO, placebo; W, week.
Impact of Prior Anti-TNF-α Therapy on Treatment Effects
Anti-TNF-α experienced vs anti-TNF-α naïve at baseline
With the exception of disease duration, there was no difference in baseline patient characteristics between patients experienced or naïve to anti-TNF-α (Supplementary Table 3). There were also no significant differences in baseline levels of serum analytes between the group experienced to anti-TNF-α and the group naïve to anti-TNF-α (Supplementary Figure 3). Spearman’s Rank correlation coefficients with SES-CD were similar between the 2 subgroups at baseline for most serum analytes (Figure 6A).
Figure 6.
Treatment differences on serum biomarkers between the group experienced to anti-TNF-α vs the group naïve to anti-TNF-α (mixed effects model). A, Correlation of anti-TNF-α experienced and naïve patients with SES-CD from BL over the study period. B, Log2 (fold change from BL) was determined by comparing levels at the post-treatment visit to BL and displayed as a heatmap for endoscopic responders and nonresponders in FIL and PBO treatment arms for the patients’ subgroups. Statistical significance of fold change from BL was determined using mixed effects model and corrected for multiple testing using Benjamini-Hochberg method. Abbreviations: BL, baseline; Corr, correlation; FC, fold-change; FDR, false-discovery rate; FIL, filgotinib; PBO, placebo; SES-CD, Simple Endoscopic Score for Crohn’s Disease; TNF-α, tumor necrosis factor-alpha; W, week.
Treatment differences on serum analytes in anti-TNF-α treatment subgroups
Among the biomarkers tested, there were notable differences in the kinetics with which changes in level of individual circulating biomarkers occurred (Figure 6B and Supplementary Figure 1D). Among patients that experienced endoscopic response on filgotinib treatment, many of the circulating biomarkers were reduced beginning at week 2 for the group naïve to anti-TNF-α , whereas there was gradual or delayed reductions for the group experienced to anti-TNF-α. Specifically, statistically significant reductions in CRP, SAA, serum CALP, VEGF-A, IL-6, and OSM at week 2 were observed in the naïve group, but a significant reduction in CRP was observed at week 2 in the experienced group. Among filgotinib responders that were experienced to anti-TNF-α, significant reductions at week 10 from baseline were observed for CRP, VEGF-A, IL-6, OSM, C3M, C4M, IL-17A, and IFN-γ. With the exception of C3M at week 4 in patients experienced to anti-TNF-α, no statistically significant reductions in biomarker levels were observed in the filgotinib nonresponder group. In the placebo responder group, there were significant reductions in CRP, SAA, and IL-6 levels in patients experienced to anti-TNF-α. There were no significant changes in biomarker levels in the placebo nonresponder group.
Effect of filgotinib treatment on whole blood pathways in anti-TNF-α treatment subgroups
Filgotinib treatment resulted in a reduction in activity of most immune pathways in both anti-TNF-α experienced responders and anti-TNF-α naïve responders at week 10 (Supplementary Figure 4). This included significant decreases in “IL6/JAK/STAT3,” “IFN-α response,” and “IFN-γ response” pathways for the responders who had experienced anti-TNF-α in the past and in the “IL6/JAK/STAT3” and “inflammatory response” pathways in the responders who were naïve to anti-TNF-α.
Discussion
Although advances in our understanding of the molecular mechanisms and inflammatory pathways underlying CD have been made in recent years, there remains a need for identification of biomarkers that accurately reflect disease activity.7 The ideal biomarker is less invasive than endoscopy, has high sensitivity and specificity, is readily available, and is cost-effective.5,7 However, it should be kept in mind that circulating biomarkers do not provide direct measurements from the site of the disease, in this case the intestinal mucosa. Therefore, they should be correlated to measures that relate directly to the tissue-level disease activity.
The present study found that of the 31 serum analytes examined, correlations were found only with 2 of the 5 clinical metrics, SES-CD and FC, at a moderate level. This is of note, as SES-CD and FC are clinically objective markers of CD; SES-CD is an endoscopic assessment of response to therapy, whereas FC is a marker of inflammation of the intestinal mucosa.4,5 The other clinical metrics used—CDAI and its subcomponents SFS and APS—are subjective measures of CD activity and had little or no correlation with serum analytes (Figure 1).
The analytes most highly correlated to both SES-CD and FC included the acute phase proteins CRP and SAA. C-reactive protein is a nonspecific marker of systemic inflammation, and SAA is a marker of general inflammation or infection that has been found to be positively associated with active endoscopic and histologic inflammation in CD.5,31 The positive association between CRP and FC corroborates the findings of other studies.32 Additionally, the cytokines IL-6, IL-22, and OSM were correlated at a similar level. Oncostatin M is thought to be the most transcriptionally upregulated cytokine in the inflamed intestinal mucosa of patients with CD; IL-6 signaling is potentially involved in the development of CD, whereas IL-22 is important for epithelial homeostasis in the intestines.33–35 The moderate level of correlation between these individual circulating biomarkers and endoscopic disease activity suggests that they are not likely to bring added diagnostic utility when used in isolation, but rather, could bring value as part of composite biomarker panel operating in a pattern or signature.5 Further studies will be required to establish such a panel with robust performance characteristics.
Our data also show that the type 3 and 4 collagen degradation markers, C3M and C4M, had a moderate correlation with FC and a weak correlation with SES-CD (Figure 1). Chronic inflammation, as associated with CD, leads to excessive extracellular matrix (ECM) remodeling, releasing protein fragments into the circulation where they have potential to be used as biomarkers.36 In a study by Mortensen et al, it was demonstrated that ECM remodeling was different between ulcerative colitis (UC) and CD and that the best combination of markers to use to make a differentiation between the 2 was VICM, C3M, and C4M, all of which gave high diagnostic accuracy.36
It is thought that the innate immune response plays an important role in the development of CD, with secreted cytokines triggering and differentiating many T cells.14 Crohn’s disease may also involve an imbalance of regulatory T, T helper (Th)-1, and Th17 cells in the intestinal mucosa.14, 32 A positive correlation between multiple Th1- and Th17-associated cytokines and FC levels has been observed.32 However, in this study, apart from IL-6, none of the analytes that are known to be involved in this imbalance, for example IFN-γ, were correlated with SES-CD or FC at baseline.14
As no single biomarker has demonstrated the necessary sensitivity and specificity to act alone to evaluate CD, the use of whole blood mRNA gene expression and pathway activities might prove to be of benefit. Gene expression profiles have been used to differentiate between active and inactive CD and between CD and UC.5 We found that gene expression mapping into Hallmark pathways showed moderate correlation between the SES-CD and FC objective clinical metrics, but weak or no correlation with the subjective CDAI metrics. The “WNT beta catenin signaling,” which is a key regulatory pathway in the intestinal mucosa maintaining epithelial homeostasis, had a negative moderate correlation with SES-CD.37 As might be expected, the pathways that had a positive moderate correlation with FC were “inflammatory response” and “reactive oxygen species.”
In the FITZROY study, filgotinib induced clinical remission in more (47%) patients with CD than in those given placebo (23%), providing some evidence that inhibition of the JAK1-STAT pathway can lead to endoscopic improvement in CD.15 With these results in mind, the present study determined that there was a statistically significant reduction of a large number of serum analytes in those patients who endoscopically responded to filgotinib over the study period, which would appear to reflect the treatment effect. In addition, filgotinib treatment reduced all serum analytes that had at least a moderate positive correlation with SES-CD at baseline, which would be in alignment with the previous finding. The effect of filgotinib on whole blood pathway activity scores in responders was noted in the systemic inflammation pathways (such as “IL6/JAK/STAT3”), as might be expected. Although SES-CD and FC were most correlated with changes in blood biomarkers and transcriptional pathways in our analysis, it should be noted that the phase 2 clinical trial did not demonstrate statistically significant differences in improvement in SES-CD or FC between filgotinib and placebo, possibly due to low sample size of the placebo group. Overall, the reduction was far more marked in the filgotinib responders than the nonresponders (Table 1 and Supplementary Figure 5). This is in keeping with the fact that by week 10, the SES-CD measurement, which was significantly reduced in responders by week 10, should reflect the treatment effect of filgotinib and is consistent with the clinical findings.
Table 1.
Summary of peripheral biomarkers that are associated with disease severity at baseline or endoscopic improvement with filgotinib treatment.
| Functional Category | Peripheral Biomarkers | Reflect Endoscopic Disease Severity at Baseline | Filgotinib treatment | |
|---|---|---|---|---|
| Kinetics | Responder vs Nonresponder | |||
| Systemic inflammation | CRP, SAA | Moderate | Early and sustained decrease | Greater in respondersa |
| OSM, IL-6 | Moderate | Early and sustained decrease | Only in responders | |
| IL6_JAK_STAT3_SIGNALLING INFLAMMATORY_RESPONSE | Moderate | Decrease at week 10 (no info on early time) | Only in responders | |
| ECM turnover epithelial homeostasis | C3M, C4M, IL-22 | Moderate | Delayed decrease (week 4) | Greater in respondersa |
| Pro-inflammatory (Th1/17) | IFNg, IL-17A | No | Delayed decrease (week 10) | Only in responders |
| INTERFERON_GAMMA_RESPONSE | No | Decrease at week 10 | Only in responders | |
| Anti-inflammatory (Treg) | IL-10, TGFb1 | No | Early decrease, return to BL | Only in responders |
Denotes that both responders and nonresponders had a statistically significant change, but the magnitude of the change was larger for the responders.
Although we observed a reduction in circulating levels of IL-6, OSM, and IL-10, which signal through JAK1, the changes in these cytokine levels in patients treated with filgotinib likely reflect indirect effects. The sustained reduction in pro-inflammatory cytokines IL-6 and OSM and the transient reduction in anti-inflammatory IL-10 may indicate an overall restoration toward homeostasis in the gut. Because these markers correlate with endoscopic findings at baseline, their reduction upon treatment with filgotinib is suggestive of their potential utility as biomarkers of CD activity. Because they showed an early differential reduction between patients with CD, with and without clinical benefit after filgotinib, they could potentially be early predictors of this response. Logistic regression revealed that both IL-6 and IL-10 were prognostic for endoscopic response, although neither was predictive.
Given that the gold-standard treatment therapy for CD is biologic therapy with TNF blockers, it was of interest to note the association between prior anti-TNF-α treatment on biomarker changes in this study. At baseline, there was no difference between the levels of serum analytes between the experienced and naïve groups, including SES-CD correlation. However, there were notable differences in the kinetics of changes in levels of the measured analytes within the 10-week treatment period, with many of the circulating biomarkers found to be reduced earlier in patients that experienced endoscopic response on filgotinib treatment without previous anti-TNF-α use.
The present study has identified a number of correlates as noninvasive biomarkers of disease activity, which would need to be verified in a larger study. It must be acknowledged that none of these biomarkers is highly correlated on its own and would likely be best used as part of a composite measure. For example, combination of serum IL-6, OSM, or IL-22 levels with FC measurement might augment the level of correlation with disease activity.
The study has several limitations. Due to the very nature of surrogate biomarkers (ie, their location in the peripheral circulation), there were no measurements of cytokines or genes/pathways conducted in the actual tissue of disease, the intestinal mucosa. If analytes with potential as CD biomarkers could also be tested at the site of disease, then the findings could be compared with those from the peripheral circulation for verification and clinically meaningful surrogate biomarkers established. A further limitation was that no endoscopic data were taken at weeks 2 and 4, so we lacked direct comparison at these time points; SES-CD scores were available only at baseline and week 10. It must also be noted that an endoscopic response is binary in nature, and so improvement in one patient might still reflect a worse level of disease in another. Finally, the statistical analyses of biomarker changes in filgotinib vs placebo responders was constrained by the limited number of placebo endoscopic responders, whereas the patient distribution between the filgotinib and placebo groups was also uneven (biomarker analysis set: filgotinib n = 126; placebo n = 42).
Conclusions
The present study identified several circulating factors (CRP, SAA, OSM, IL-22, IL-6, C3M, and C4M) that have potential use as biomarkers to assess CD activity. Further, the levels of many of these candidate biomarkers were significantly reduced upon treatment with filgotinib, particularly in patients with CD that achieved clinical benefit from this treatment. The utility of these biomarkers will need additional exploration in larger clinical studies to define how they might best be used as surrogate markers of disease activity or response to treatment.
Supplementary Material
Acknowledgments
The authors wish to thank Lesley Blogg, PhD, Bryan Thibodeau, PhD, and Claudette Knight, PharmD, of Fishawack Health for medical writing assistance in the preparation of this article, which was funded by Gilead Sciences, Inc.
Contributor Information
Xavier Roblin, Gastroenterology Unit, University Hospital of Saint Etienne, Saint-Priest-en-Jarez, France.
Adrian Serone, Gilead Sciences, Inc., Foster City, CA, USA.
Oh Kyu Yoon, Gilead Sciences, Inc., Foster City, CA, USA.
Luting Zhuo,, Gilead Sciences, Inc., Foster City, CA, USA.
Ethan Grant, Gilead Sciences, Inc., Foster City, CA, USA.
Jacky Woo, Gilead Sciences, Inc., Foster City, CA, USA.
Jinfeng Liu, Gilead Sciences, Inc., Foster City, CA, USA.
René Galien, Galapagos SASU, Romainville, France.
Geert D’Haens, Inflammatory Bowel Disease Centre, Academic Medical Centre, 1105 AZ Amsterdam, Netherlands.
Funding
This study was funded by Gilead Sciences, Inc.
Conflicts of Interest
X.R. reports relationships with AbbVie, MSD, Janssen Cilag, and Takeda. A.S., O.K.Y., L.Z., E.G., and J.L. are employees and stockholders of Gilead Sciences, Inc. J.W. was an employee of Gilead Sciences, Inc. at the time of study conduct. R.G. is an employee of Galapagos SASU and holds shares of Galapagos. G.D.H. has served as advisor for AbbVie, Ablynx, Allergan, Alphabiomics, Amakem, Amgen, AM Pharma, Applied Molecular Therapeutics, Arena Pharmaceuticals, AstraZeneca, Avaxia, Biogen, Bristol-Meyers Squibb, Boehringer Ingelheim, Celgene/Receptos, Celltrion, Cosmo, DSM Pharma, Echo Pharmaceuticals, Eli Lilly, Engene, Exeliom Biosciences, Ferring, DrFALK Pharma, Galapagos, Genentech/Roche, Gilead Sciences Inc. GlaxoSmithKline, Gossamerbio, Hospira/Pfizer, Immunic, Johnson and Johnson, Kintai Therapeutics, Lycera, Medimetrics, Millenium/Takeda, Medtronics, Mitsubishi Pharma, Merck Sharp & Dome Corp., Mundipharma, Nextbiotics, Novo Nordisk, Otsuka, Pfizer/Hospira, Photopill, ProciseDx, Prodigest, Prometheus laboratories/Nestle, Progenity, Protagonist, RedHill, Robarts Clinical Trials, Salix, Samsung Bioepis, Sandoz, Seres/Nestle, Setpoint, Shire, Teva, Tigenix, Tillotts, Topivert, Versant, and Vifor.
References
- 1. Feuerstein JD, Cheifetz AS.. Crohn disease: epidemiology, diagnosis, and management. Mayo Clin Proc. 2017;92:1088–1103. [DOI] [PubMed] [Google Scholar]
- 2. Boyapati R, Satsangi J, Ho GT.. Pathogenesis of Crohn’s disease. F1000prime Rep. 2015;7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lillis C. Can Crohn’s disease be fatal? Medical News Today. 2018. Accessed Jun 4, 2020. https://www.medicalnewstoday.com/articles/323438
- 4. Moein S, Qujeq D, Vaghari Tabari M, et al. Diagnostic accuracy of fecal calprotectin in assessing the severity of inflammatory bowel disease: from laboratory to clinic. Caspian J Intern Med. 2017;8:178–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barnes EL, Burakoff R.. New biomarkers for diagnosing inflammatory bowel disease and assessing treatment outcomes. Inflamm Bowel Dis. 2016;22:2956–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bourgonje AR, von Martels JZH, Gabriëls RY, et al. A combined set of four serum inflammatory biomarkers reliably predicts endoscopic disease activity in inflammatory bowel disease. Front Med (Lausanne). 2019;6:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Norouzinia M, Chaleshi V, Alizadeh AHM, Zali MR.. Biomarkers in inflammatory bowel diseases: insight into diagnosis, prognosis and treatment. Gastroenterol Hepatol Bed Bench. 2017;10:155–167. [PMC free article] [PubMed] [Google Scholar]
- 8. de Silva S, Devlin S, Panaccione R.. Optimizing the safety of biologic therapy for IBD. Nat Rev Gastroenterol Hepatol. 2010;7:93–101. [DOI] [PubMed] [Google Scholar]
- 9. Adegbola SO, Sahnan K, Warusavitarne J, et al. Anti-TNF therapy in Crohn’s disease. Int J Mol Sci. 2018;19:2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Samaan M, Campbell S, Cunningham G, et al. Biologic therapies for Crohn’s disease: optimising the old and maximising the new. F1000Res. 2019;8:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pagnini C, Pizarro TT, Cominelli F.. Novel pharmacological therapy in inflammatory bowel diseases: beyond anti-tumor necrosis factor. Front Pharmacol. 2019;10:671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colombel JF, Panaccione R, Bossuyt P, et al. Effect of tight control management on Crohn’s disease (CALM): a multicentre, randomised, controlled phase 3 trial. Lancet. 2017;390:2779–2789. [DOI] [PubMed] [Google Scholar]
- 13. Loddo I, Romano C.. Inflammatory bowel disease: genetics, epigenetics, and pathogenesis. Front Immunol. 2015;6:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sanchez-Munoz F, Dominguez-Lopez A, Yamamoto-Furusho JK.. Role of cytokines in inflammatory bowel disease. World J Gastroenterol. 2008;14:4280–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vermeire S, Schreiber S, Petryka R, et al. Clinical remission in patients with moderate-to-severe Crohn’s disease treated with filgotinib (the FITZROY study): results from a phase 2, double-blind, randomised, placebo-controlled trial. Lancet. 2017;389:266–275. [DOI] [PubMed] [Google Scholar]
- 16. Ma C, Jairath V, Vande Casteele N.. Pharmacology, efficacy and safety of JAK inhibitors in Crohn’s disease. Best Pract Res Clin Gastroenterol. 2019;38-39:101606. [DOI] [PubMed] [Google Scholar]
- 17. Shuai K. Regulation of cytokine signaling pathways by PIAS proteins. Cell Res. 2006;16:196–202. [DOI] [PubMed] [Google Scholar]
- 18. Babon JJ, Lucet IS, Murphy JM, et al. The molecular regulation of Janus kinase (JAK) activation. Biochem J. 2014;462:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghoreschi K, Laurence A, O’Shea JJ.. Janus kinases in immune cell signaling. Immunol Rev. 2009;228:273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Van Rompaey L, Galien R, van der Aar EM, et al. Preclinical characterization of GLPG0634, a selective inhibitor of JAK1, for the treatment of inflammatory diseases. J Immunol. 2013;191:3568–3577. [DOI] [PubMed] [Google Scholar]
- 21. World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. [DOI] [PubMed] [Google Scholar]
- 22.The International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use. Integrated Addendum to ICH E6(R1): Guideline For Good Clinical Practice E6(R2). Accessed Jun 4, 2020. https://database.ich.org/sites/default/files/E6_R2_Addendum.pdf
- 23. Patro R, Duggal G, Love MI, et al. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liberzon A, Birger C, Thorvaldsdóttir H, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Best WR, Becktel JM, Singleton JW, Kern F Jr. Development of a Crohn’s disease activity index. National cooperative Crohn’s disease study. Gastroenterology. 1976;70:439–444. [PubMed] [Google Scholar]
- 28. Benjamini Y, Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B (Methodological). 1995;57:289–300. [Google Scholar]
- 29. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Law CW, Chen Y, Shi W, Smyth GK.. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yarur AJ, Quintero MA, Jain A, et al. Serum amyloid A as a surrogate marker for mucosal and histologic inflammation in patients with Crohn’s disease. Inflamm Bowel Dis. 2017;23:158–164. [DOI] [PubMed] [Google Scholar]
- 32. Bourgonje AR, von Martels JZH, de Vos P, et al. Increased fecal calprotectin levels in Crohn’s disease correlate with elevated serum Th1- and Th17-associated cytokines. Plos One. 2018;13:e0193202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mizoguchi A. Healing of intestinal inflammation by IL-22. Inflamm Bowel Dis. 2012;18:1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim WM, Kaser A, Blumberg RS.. A role for oncostatin M in inflammatory bowel disease. Nat Med. 2017;23:535–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu G, Jin S, Jiang Q.. Interleukin-6 receptor and inflammatory bowel disease: a Mendelian randomization study. Gastroenterology. 2019;156:823–824. [DOI] [PubMed] [Google Scholar]
- 36. Mortensen JH, Godskesen LE, Jensen MD, et al. Fragments of citrullinated and MMP-degraded vimentin and MMP-degraded type III collagen are novel serological biomarkers to differentiate Crohn’s disease from ulcerative colitis. J Crohns Colitis. 2015;9:863–872. [DOI] [PubMed] [Google Scholar]
- 37. Moparthi L, Koch S.. WNT signaling in intestinal inflammation. Differentiation. 2019;108:24–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.