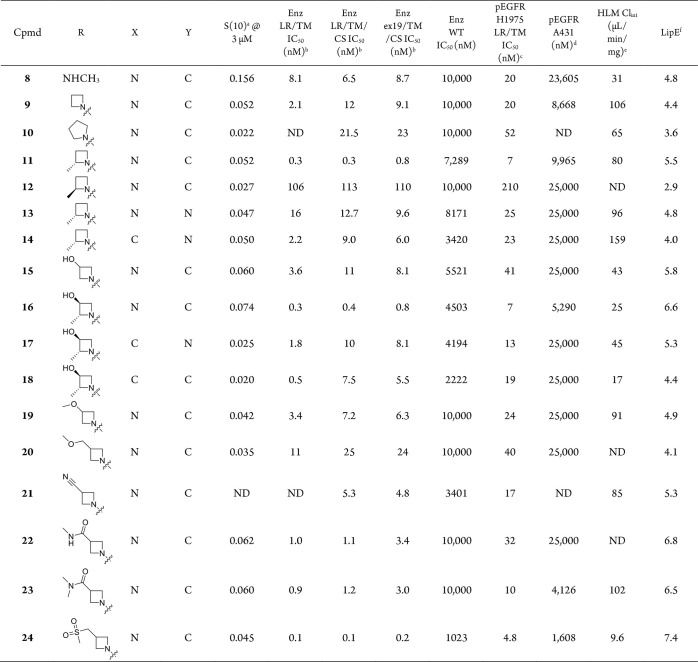
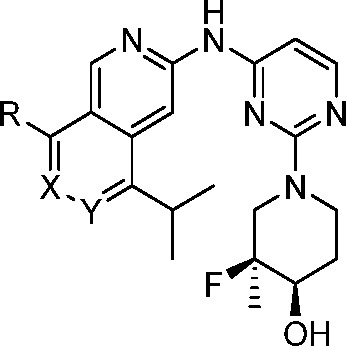
Table 2. Structure–Activity Relationship of Selected Analogs of Compound 6.
DiscoverX’s KINOMEscan selectivity profiling at 3 μM, S(10) = (number of non-mutant kinases with %Ctrl < 10)/(number of non-mutant kinases tested).
Biochemical assays using different EGFR variants measure inhibition in the presence of 1 mM ATP, and compounds were incubated with enzymes for 10 min before ATP and peptide substrate were added (for more details see Experimental Section). EGFR LR/TM = EGFR L858R/T790M, EGFR LR/TM/CS = L858R/T790M/C790S, and ex19/TM/CS = ex19del(746–750)/L858R/C790S.
H1975 is a human lung cancer cell line harboring the EGFR L858R/T790M mutation.
A431 is a cell line in which EGFR is amplified.
Intrinsic clearance obtained from isolated human liver microsomes.
LipE = −log(LRTMCS Enz IC50) – log D (calculated from ACD labs @ pH = 7.4). ND: not determined.