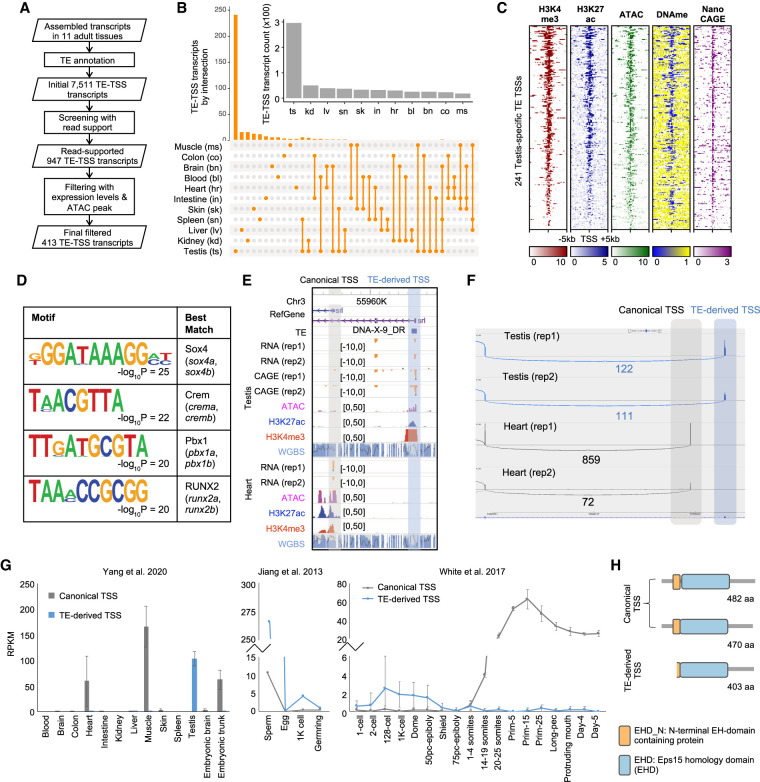
Figure 5.
Tissue-specific alternative promoters derived from TEs. (A) Flowchart describing the methods used to identify TE-TSS transcripts. (B) UpSet plot of TE-TSS transcripts expressed in zebrafish tissue. (C) Heat maps of ChIP-seq, ATAC-seq signals, DNA methylation levels (DNAme), and nanoCAGE over 10-kb regions centered on testis-specific TE-derived TSSs. (D) Table of motifs enriched in testis-specific TE-derived alternative promoters, and the best TF matches. (E) Epigenome Browser view of TE-derived srl canonical and alternative promoters. (F) Sashimi plot showing RNA-seq reads spanning exon-exon junctions. Only reads anchored on the canonical exon 2 are shown for simplicity. (G) Plots of canonical and TE-derived TSS usages for srl in different tissues and developmental stages. (H) Protein structures from transcripts initiated from canonical TSS and putative protein structures from the TE-TSS transcript srl.