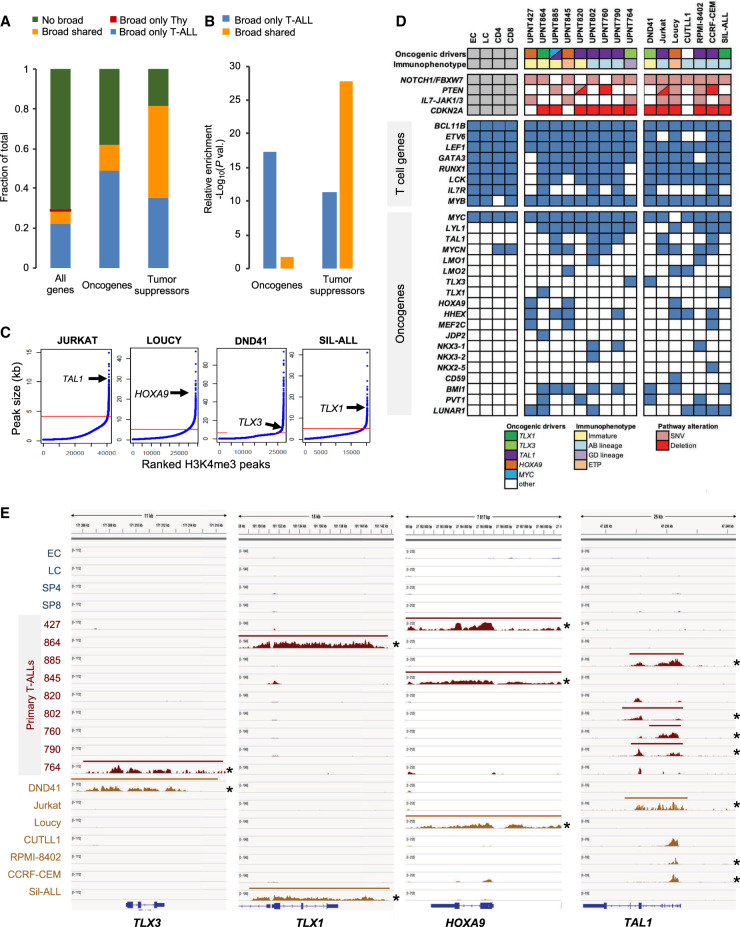
Figure 2.
Characterization of broad H3K4me3 domains in T-ALL samples. (A) Distribution of the indicated category of genes associated with broad H3K4me3 domains only in thymocytes (Tc), only in T-ALL, in both thymocytes and T-ALL (shared), or without broad H3K4me3 domain (no broad). Oncogenes and tumor suppressors are listed in Supplemental Table S5. Note that no oncogene or tumor suppressor gene was associated with a broad H3K4me3 domain only in thymocytes. (B) Relative enrichment for oncogenes and tumor suppressors associated with the broad H3K4me3 domain only in T-ALL or shared between T-ALL and thymocytes was assessed by hypergeometric tests. (C) The H3K4me3 peaks were ranked according to their size for the indicated T-ALL cell lines. The inflection point allowing the selection of broad H3K4me3 domains is indicated by a red line. The position of the broad H3K4me3 domains associated with the representative oncogene is shown for each cell line. (D) Table showing the association of broad H3K4me3 domains with oncogenes frequently activated in T-ALL. The maturation states and main oncogenetic alterations of T-ALL samples are shown. (E) Examples of key oncogenes associated with broad H3K4me3 domains in T-ALL cells. H3K4me3 ChIP-seq signal and broad H3K4me3 domain tracks are shown for each thymocyte population and T-ALL sample. Asterisk indicates ectopic expression of the oncogene.