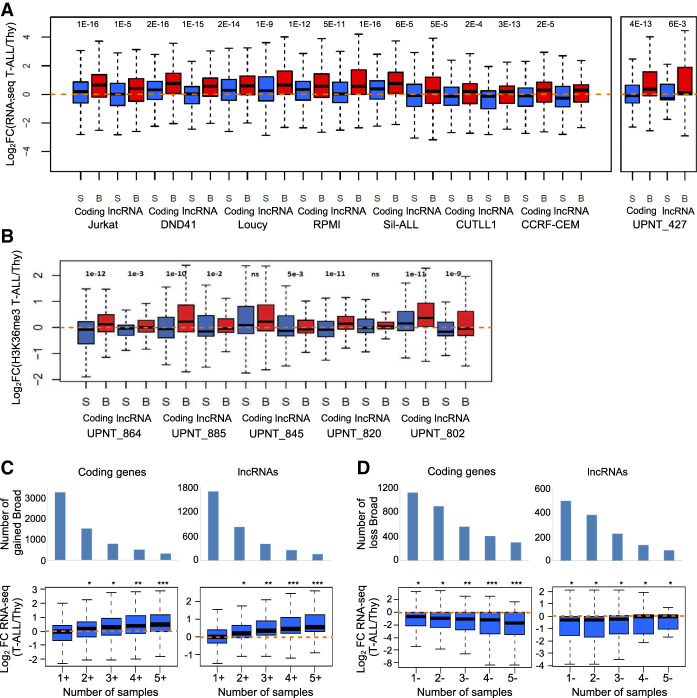
Figure 4.
Dynamic expression of broad H3K4me3 domain–associated genes. (A,B) Two sets of equally expressed genes associated with broad (B) or sharp H3K4me3 peak (S) in the indicated T-ALL sample were defined based on RNA-seq data (A) or H3K4me3 gene-body enrichment (B). The fold change in expression between the T-ALL samples and the normal T cell populations was determined. (C,D) Gene expression in the function of the frequency of association with broad H3K4me3 domains in primary T-ALL samples. Genes specifically associated with broad H3K4me3 domains in T-ALL samples were separated according to the cumulative number of samples with gain (C) or loss (D). The fold change expression of coding genes and lncRNAs between the thymus and a series of independent 41 T-ALLs was analyzed. (***) P < 0.001, (**) P < 0.01, (*) P < 0.05, Wilcoxon rank-sum test; (ns) not significant.