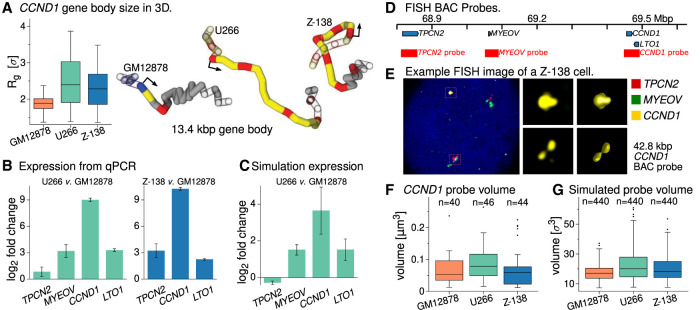
Figure 6.
Gene size and expression measurements. (A, left) Box plot showing the distribution of the radius of gyration of the CCND1 gene body (a measure of its 3D size) in simulations for each of the three cell types (440 configurations for each case). (Right) Typical snapshots of the gene and a 5-kbp flanking region on each side. The gene body is shown as a solid tube, with flanking regions shown as an outline. The arrow shows the transcription start site. The polymer is colored according to the simulation input data: Red indicates active protein binding sites (DHSs); blue, polycomb protein binding sites; and yellow, open chromatin (H3K27ac). (B, left) Bar plot showing the change in expression level in U266 cells compared to GM12878 obtained from qPCR measurements (see Supplemental Methods Section 6 and Supplemental Fig. S3 for further details). The bar height gives the log2 fold change based on an average over three replicates; the error bar shows the standard error of the mean. (Right) Similar plot for Z-138 cells compared to GM12878. No MYEOV expression was detected in these cells. (C) Bar plot showing the fold change in expression level in U266 compared to GM12878 based on the expression prediction from simulations. As detailed in the text, we do not expect a linear mapping between the predicted and real expression levels, so one can only compare qualitatively with the qPCR. Nevertheless, this shows the same trend across the genes as in panel B. See Supplemental Methods for further details. (D) Map of the CCND1 locus (hg19) showing the positions of the fosmid probes used in FISH experiments. (E) Example FISH images. The left image shows a whole cell with red, green, and yellow probes covering different genes as indicated. Middle images show the yellow probe spots only (regions indicated by orange boxes in the whole-cell image). Right images show a 3D reconstruction of the gold probes as used to measure the volume (see Supplemental Methods for details). (F) Box plot showing the distributions of the volume of the probe covering CCND1 in the three cell lines (see text and Supplemental Methods for details). The number of probe spots measured in each case (n) is indicated. See Supplemental Figure S10 for similar plots for the other probes. (G) Box plot showing a similar measurement but extracted from n = 440 simulated configurations from each cell line (see Supplemental Methods for details of the calculation; simulation length units roughly map to physical units as σ ≈ 21.8 nm, so σ3 ≈ 1.04 × 10−5 µm).