Figure 1.
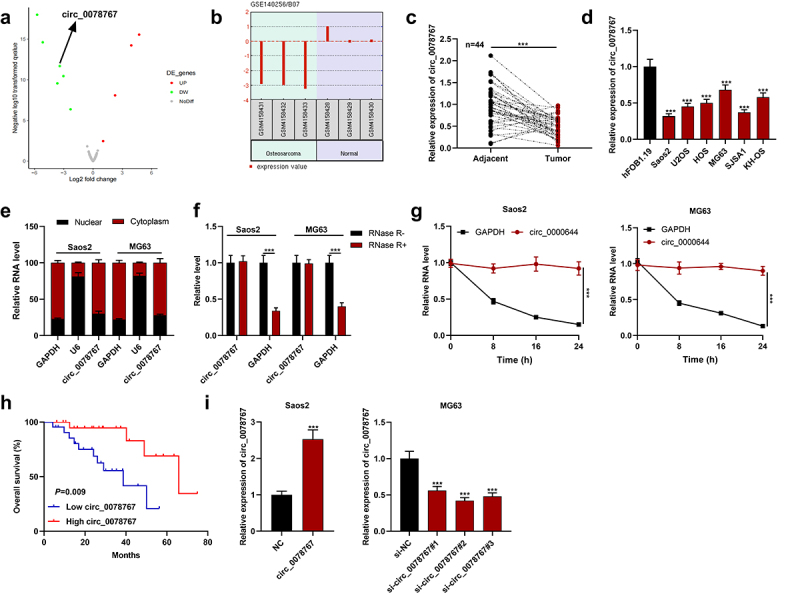
Circ_0078767 is downregulated in OS cells and tissues.
(a). The volcano plot was used to show the differentially expressed circRNAs in OS tissues compared with normal tissues in the GSE140256 dataset. CircRNAs with significantly up-regulated expression were marked in red, and the down-regulated circRNAs were marked in green. Grey represented circRNAs that were not significantly differentially expressed. Screening criteria: log2(fold change) > 1 or < −1 and P < 0.05.(b). The expression profile of circ_0078767 in OS tissue (n = 3) and normal adjacent tissue (n = 3).(c&d). Detection of circ_0078767 expression in OS tissues and cells by qRT-PCR.(e). Subcellular distribution analysis of circ_0078767 in Saos2 and MG63 cells.(f). The relative levels of circ_0078767 and GAPDH mRNA in Saos2 and MG63 cells with or without RNase R incubation were detected by qRT-PCR.(g). Saos2 and MG63 cells were treated with actinomycin D, and the relative expression levels of circ_0078767 and GAPDH were examined by qRT-PCR at different times (0, 8, 16 and 24 h).(h). Patients with OS were divided into two groups according to the median level of circ_0078767 expression. Overall survival was analyzed utilizing the Kaplan-Meier method with the log-rank test.(i). Detection via qRT-PCR of circ_0078767 expression in Saos2 cells transfected with circ_0078767 overexpression plasmids and MG63 cells transfected with circ_0078767 siRNAs.All experiments were carried out in triplicate. ***P < 0.001.
