Figure 2.
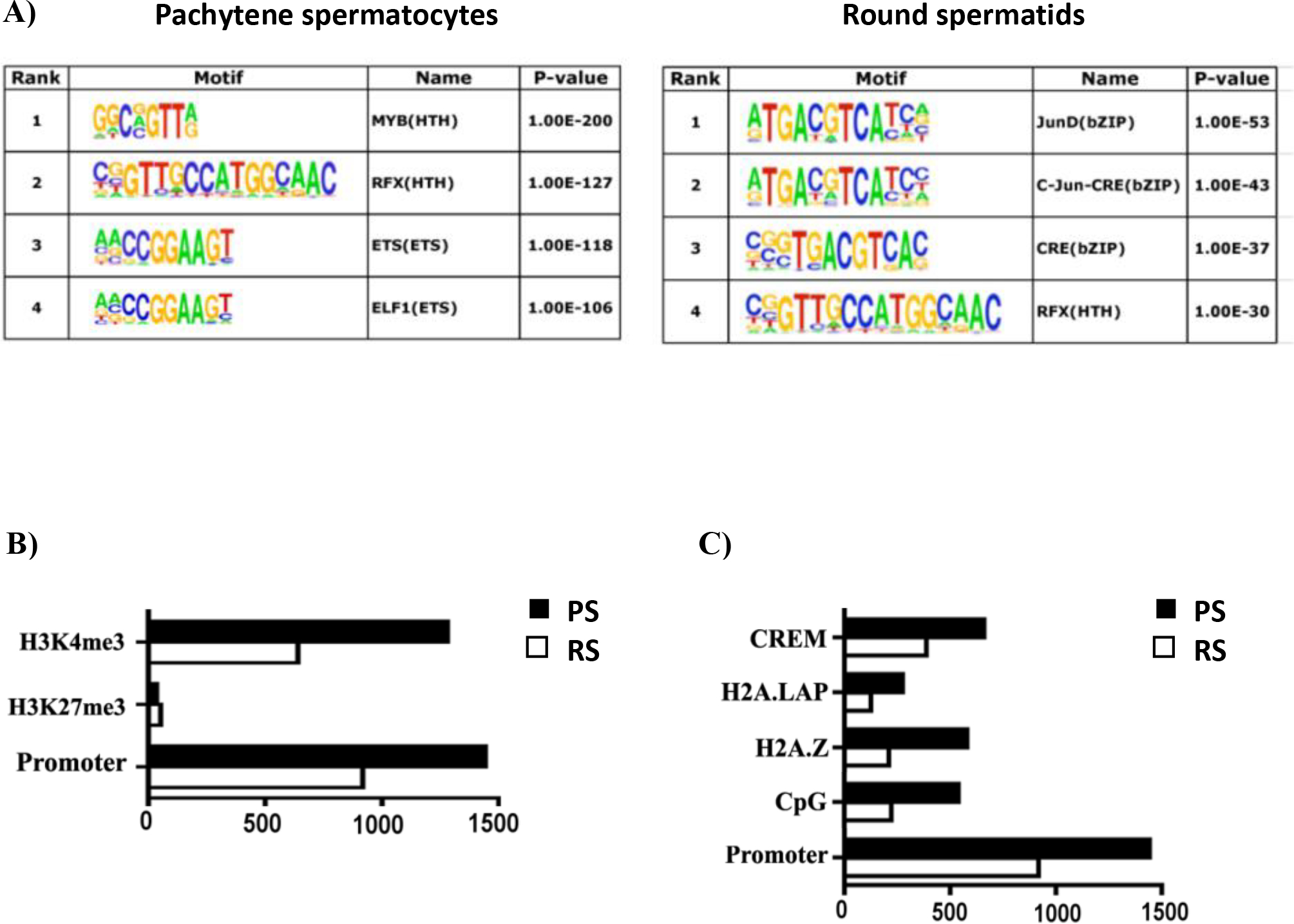
Characterization of BRDT binding site occupancy in pachytene spermatocytes (PS) and round spermatids (RS). (A) BRDT in vivo consensus binding site predicted by MEME and presented as a sequence LOGO. A total of 500 peak regions were analyzed for overrepresented motifs. Enriched transcription factor motifs lying within BRDT binding sites in PS and RS. (B) Correlative analysis of H3K4me3, H3K27me3 with BRDT binding site occupancy. (C) Correlative analysis of CpG, H2AZ, H2A.LAP, CREM with BRDT binding site occupancy. The x-axis in (B) and (C) refers to the number of associations with BRDT binding sites.
