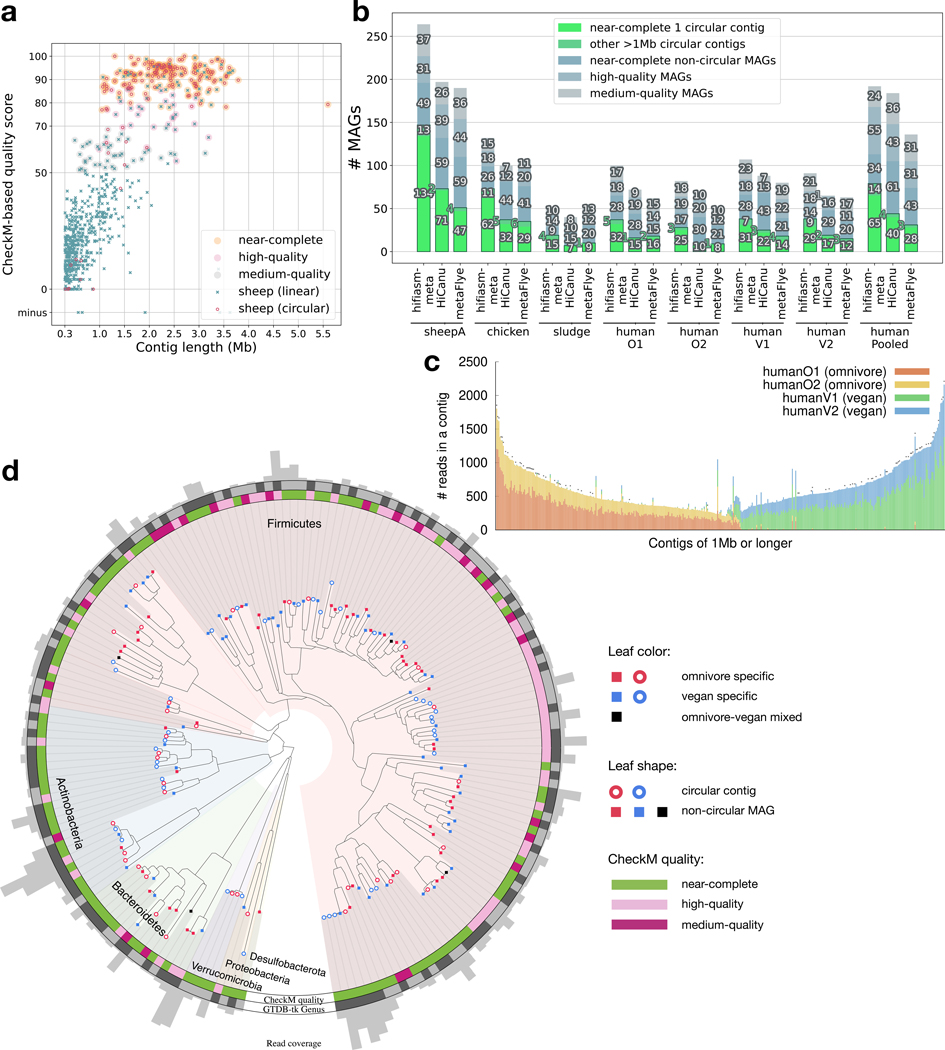
Figure 1.
Metagenome assemblies of empirical datasets. (a) Quality score of contigs longer than 300kb from the hifiasm-meta assembly of sheepA. The quality score of a MAG is defined as “completeness −5 × contamination” based on CheckM reports. (b) CheckM evaluation. A MAG is “near-complete” if its CheckM completeness is ≥90% and its contamination level ≤5%, is “high-quality” if completeness ≥70% and contamination ≤10%, or is “medium-quality” if its quality score is ≥50%. “HumanPooled” represents the co-assembly of all four human gut samples. (c) Composition of long contigs in the hifiasm-meta co-assembly of four human gut samples. Each bar represents a contig of ≥1Mb in length. It gives the number of reads used in the contig. Each color corresponds to a human gut sample. A cross at the top of a bar indicates the contig being circular. (d) Phylogeny of human gut MAGs from the co-assembly. A colored clade corresponds to a phylum inferred by GTDB-Tk18,19. A MAG is omnivore/vegan-specific if 90% of reads in the MAG come from omnivore/vegan samples, respectively. Two adjacent leaves coming in the same genus have the same shade on the “Genus” outer ring.