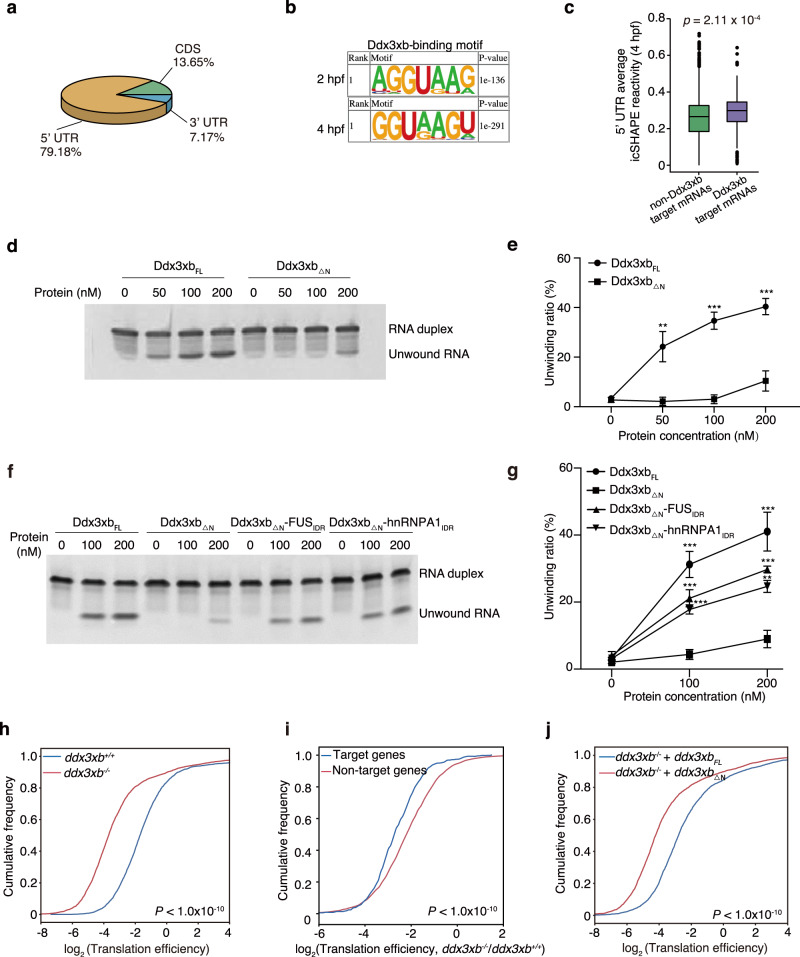
Fig. 5. Condensation of Ddx3xb regulates maternal mRNA translation via unwinding RNA structures.
a Pie chart depicting relative fraction of Ddx3xb-binding reads that mapped to 5’ UTR, CDS and 3’ UTR regions of mRNAs identified by RIP-seq. mRNA target regions were subdivided into three transcript segments. b Sequence motif identified within the binding region of Ddx3xb at 5’ UTR by HOMER. c Boxplot showing the average icSHAPE reactivity in 5’ UTR of non-Ddx3xb target mRNAs and Ddx3xb target mRNAs at 4 hpf (a high icSHAPE reactivity implies a single-stranded conformation, a low icSHAPE reactivity implies a double-stranded conformation). P values were calculated by the Kruskal Wallis test. d, e In vitro helicase assays using different doses (0 nM, 50 nM, 100 nM, and 200 nM) of purified Ddx3xbFL and Ddx3xb△N proteins. The unwinding ratio was calculated by (unwound RNA)/((unwound RNA) + (RNA duplex)). Error bars, means ± SD (n = 3). P values were determined by the two-tailed Student’s t-test. **P < 0.01, ***P < 0.001. f, g In vitro helicase assays using different doses (0 nM, 100 nM, and 200 nM) of purified Ddx3xb△N, Ddx3xb△N-hnRNPA1IDR, and Ddx3xb△N-FUSIDR proteins. The unwinding ratio was calculated by (unwound RNA)/((unwound RNA) + (RNA duplex)). Error bars, means ± SD (n = 3). P values were determined by the two-tailed Student’s t-test. **P < 0.01; ***P < 0.001. h Cumulative distribution of the translation efficiency of Ddx3xb mutant and wild-type embryos at 4 hpf. P values were calculated by the Kruskal Wallis test. i Cumulative distribution of the log2 fold changes of translation efficiency of Ddx3 target genes and Ddx3 non-target genes between Ddx3xb mutant and wild-type embryos at 4 hpf. P values were calculated by the Kruskal Wallis test. j Cumulative distribution of the translation efficiency of ddx3xbFL and ddx3xb△N mRNAs-injected ddx3xb mutant embryos at 4 hpf. P values were calculated by the Kruskal Wallis test.