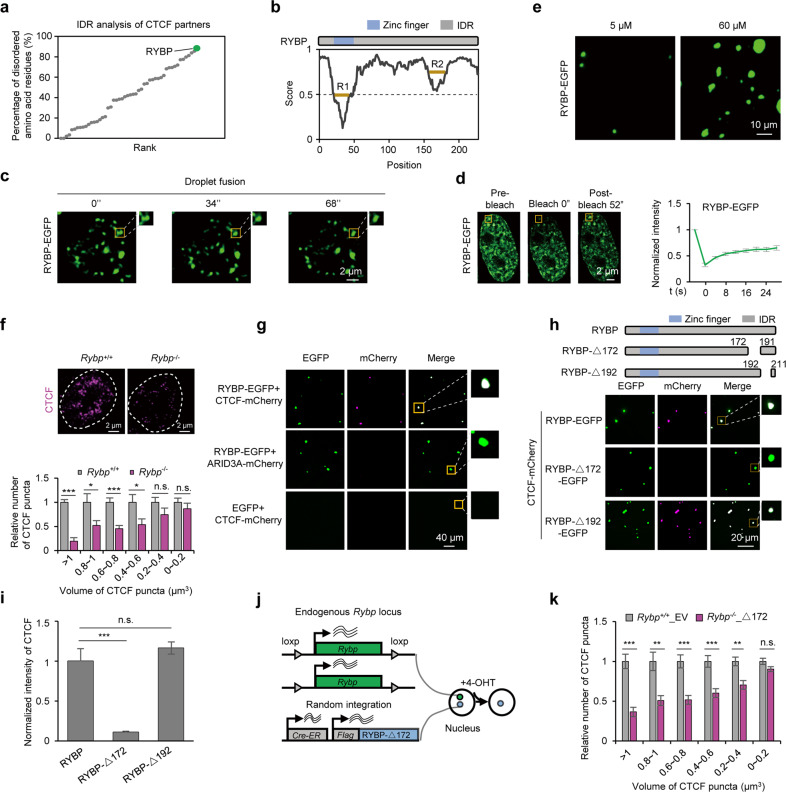
Fig. 3. RYBP facilitates CTCF to undergo phase separation.
a IDR analysis of CTCF partners, the involved interaction partners of CTCF were extracted from the BIOGRID database. Polycomb group components were also analyzed, since Polycomb body is reported to co-localize with CTCF.31 b RYBP is an intrinsically disordered protein predicted by IUPred2A.88 R1 and R2 denote the relatively low disordered regions of RYBP. c Droplet fusion behavior of RYBP puncta in an EGFP-tagged RYBP-expressing ESC (RYBP-EGFP ESC). d Representative FRAP images (left) and quantification (right) in ESCs expressing exogenous RYBP-EGFP, n = 3. The yellow box denotes the bleached puncta. e Representative images showing droplet formation of 5 µM and 60 µM RYBP-EGFP recombinant protein. f Representative immunofluorescence images (top) and statistics (bottom) of CTCF puncta before and after RYBP depletion. Welch’s t-test; Rybp+/+, n = 61 cells; Rybp–/–, n = 37 cells. P values are (from left to right): P = 7.47e–06, P = 0.0465, P = 0.0008, P = 0.0466, P = 0.1662, P = 0.3842. g RYBP droplets incorporate CTCF protein in vitro without PEG8000. The concentration of RYBP-EGFP and EGFP was 60 µM; the concentration of CTCF-mCherry and ARID3A-mCherry was 10 µM. h Top: experimental pipeline for the mutational strategy of RYBP. Bottom: full-length or mutant RYBP droplets incorporate CTCF protein without PEG8000 in vitro. The concentration of RYBP-EGFP, RYBP-Δ172-EGFP and RYBP-Δ192-EGFP was 60 µM; the concentration of CTCF-mCherry and mCherry was 10 µM. i Normalized intensity of CTCF in full-length or mutant RYBP droplets. Welch’s t-test; RYBP, n = 25; RYBP-Δ172, n = 35; RYBP-Δ192, n = 23. P values are (from left to right): P = 9.028e–06, P = 0.064. j Experimental pipeline for the construction of RYBP mutant cell line. k Relative number of different volumes of CTCF puncta in Rybp+/+_EV or Rybp–/–_Δ172 cell lines. Welch’s t-test; Rybp+/+_EV, n = 85 cells; Rybp–/–_Δ172, n = 164 cells. P values are (from left to right): P = 6.59e–05, P = 0.0032, P = 0.0003, P = 0.0007, P = 0.0015, P = 0.0736. n.s., not significant, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.