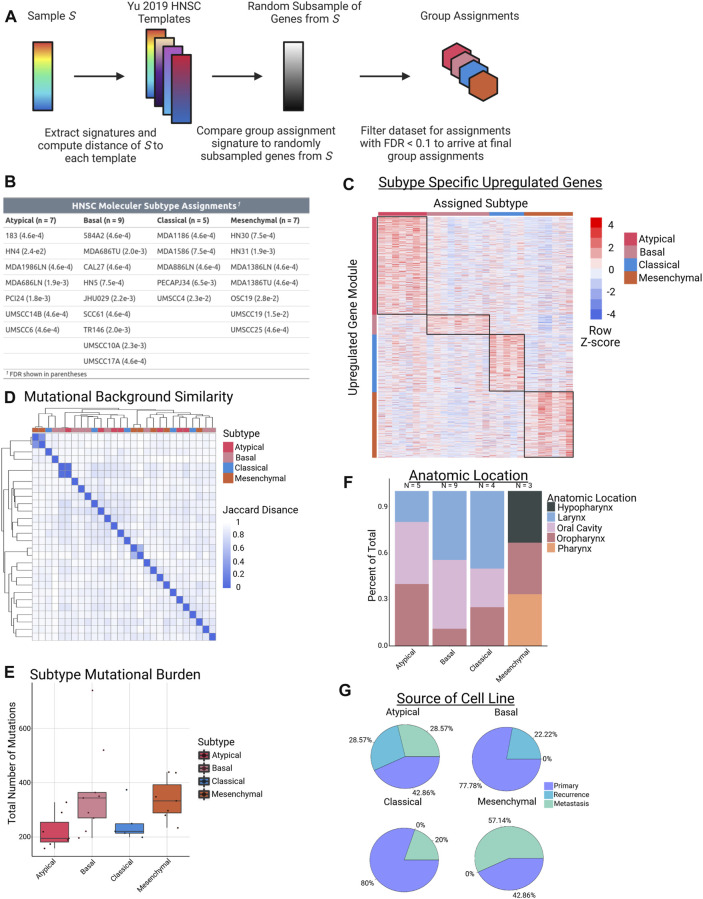
FIGURE 1.
Cell line subtype assignments and characteristics. (A) Schematic of workflow used to assign HNSCC cell lines to their respective subtypes using RNA-seq data. (B) Table of subtype assignments for each of the 28 cell lines used in this study. (C) Heatmap of gene expression modules in each molecular subtype, defined as FC > 3 in a one-vs-rest comparison. (D) Hierarchical clustering of the 28 cell lines based on Jaccard distance metrics obtained from binarized mutation counts from WES data. (E) Boxplots demonstrating total number of mutations in each sample, grouped by molecular subtype (p = NS for each comparison). (F) Stacked barplot showing distribution of cell line anatomic location for each molecular subtype. (G) Pie chart showing percentage of samples in each molecular subtype that came from primary, recurrent, or metastatic lesions.