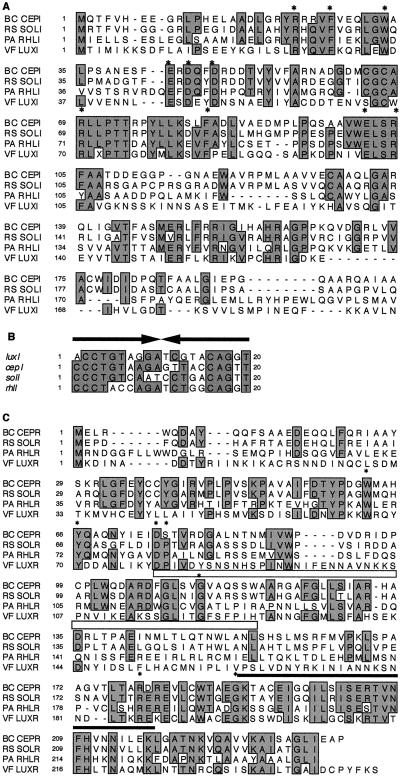
FIG. 2.
Multiple alignments of amino acid sequences from LuxIR family members and of lux box promoter elements. (A) Amino acid alignment of various LuxI family members with B. cepacia CepI (BC CEPI) generated by using the programs PC/Gene CLUSTAL and SeqVu. Boxed, shaded regions highlight amino acids conserved in at least three of the proteins. The 10 invariant amino acids characteristic of LuxI homologs are denoted with asterisks (46). Additional sequences shown are those of proteins abbreviated as follows: RS SOLI, R. solanacearum SolI (accession no. AF021840 [19]); PA RHLI, P. aeruginosa RhlI (accession no. U11811 [45]); and VF LUXI, V. fischeri LuxI (accession no. 225903 [12]). (B) Comparison of lux box sequences in the promoter regions of LuxI homologs. The sequences shown are from luxI (V. fischeri [12]), cepI (B. cepacia), solI (R. solanacearum [19]), and rhlI (P. aeruginosa [31]). The black arrows represent the inverted repeats of the palindrome sequences. Boxed, shaded regions highlight nucleotides that are identical in at least three of the sequences. (C) Amino acid alignment of various LuxR family members with B. cepacia CepR (BC CEPR) generated by using the programs PC/Gene CLUSTAL and SeqVu. Boxed, shaded regions highlight amino acids that are identical in three of the four proteins. The open bar below LuxR residues 79 to 127 represents the autoinducer binding domain (61). The solid bar above CepR residues 190 to 217 represents the putative helix-turn-helix motif that was identified by PROSITE (3). The seven invariant amino acids of LuxR homologs are denoted with asterisks (19). Additional sequences shown are those of proteins abbreviated as follows: RS SOLR, R. solanacearum SolR (accession no. AF021840 [19]); PA RHLR, P. aeruginosa RhlR (accession no. L08962 [43]); and VF LUXR, V. fischeri LuxR (accession no. 225902 [12]).