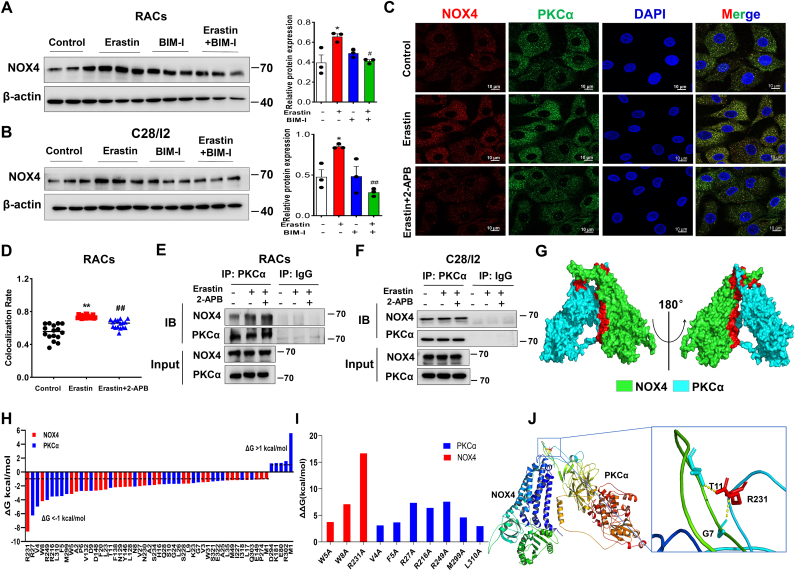
Fig. 5.
Blockade of TRPM7 suppresses PKCα-NOX4 interaction in articular chondrocytes. (A, B) Western blot analysis for NOX4 in erastin-induced RACs and C28/I2 cells treated with BIM-I (n = 3). Data are means ± SD. *P < 0.05, erastin versus control; #P < 0.05, ##P < 0.01 erastin + BIM-I versus erastin. (C) Representative photomicrographs of RACs stained to detect NOX4 (red fluorescence), PKCα (green fluorescence), and the nucleus (DAPI, blue fluorescence) treated with/without erastin (5 μM) and 2-APB (100 μM). (D) Colocalization rate of NOX4/PKCα in erastin-induced RACs treated with/without 2-APB (n = 16). Data are means ± SD. **P < 0.01 erastin versus control; ##P < 0.01 erastin + 2-APB versus erastin. (E, F) The interaction between PKCα and NOX4 in primary RACs and C28/I2 cells was analyzed by co-immunoprecipitation. (G) 3D binding structure of PKCα and NOX4 determined via molecular modeling and docking studies. (H) The energy values of NOX4 residues (R231, W8, W5) and PKCα residues (R27, V4, R249, R216, L310, F5, M299) were less than −3 kcal/mol by per-residue energy decomposition analysis. Dashed lines indicate thresholds less than −1 or greater than 1 kcal/mol. (I) Alanine scanning results showed the contributions of protein residues to the stability of the NOX4/PKCα complex. (J) Close-up view of hydrogen bonding between NOX4 residue R231 and PKCα residues (G7 and T11). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)