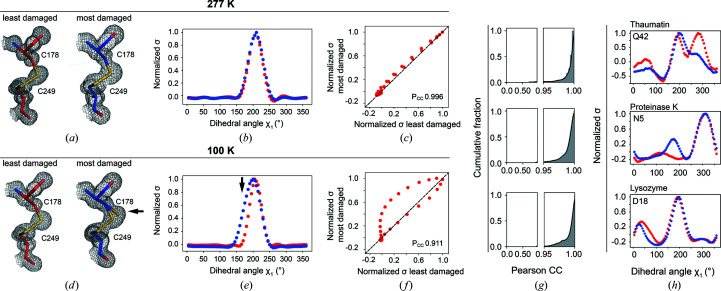
Figure 3.
Ringer analysis to quantitatively evaluate the effects of X-ray damage on side-chain rotameric distributions. The panels depict examples from the proteinase K data sets obtained in this work. Data from the least (red, left) and most (blue, right) X-ray-damaged sequential data sets collected at (a) RT (277 K) or (d) cryo temperature. Visually, it appears that there is partial X-ray-induced breakage of the disulfide bond in (d) but not in (a). The arrow points to the appearance of a new population for Cys178 that manifests as a broadened distribution. The electron density (1σ, gray mesh) and model (sticks) of the Cys178–Cys249 disulfide bond are shown. (b, e) Normalized Ringer profiles. Plots of electron density (σ) as a function of dihedral angle χ1 for Cys178 (electron density from the least and most damaged data sets is in red and blue, respectively). Each point in a Ringer profile represents the electron density for a specified dihedral angle (see Section 2). The arrow indicates a difference between the least and most damaged data sets corresponding to the appearance of a new state for Cys178 in the damaged cryo data set (d). (c, f) Correlation plots between electron-density values (σ) from (b) and (e), respectively, of the least (x axis) and most (y axis) damaged data sets. An increasing number of off-diagonal points indicates a decreasing similarity between Ringer plots. The Pearson correlation coefficient (P CC) represents the agreement between the least and most damaged Ringer profiles, with P CC = 1 for a perfect correlation (dashed line). (g) Cumulative fraction of P CC for the dihedral angle χ1 of each residue in thaumatin (top), proteinase K (middle) and lysozyme (bottom). Similar results were obtained using mean-square error analysis instead of Pearson correlation coefficients (Supplementary Fig. S6). (h) The most significant outliers with P CC ≤ 0.95 for thaumatin (top), proteinase K (middle) and lysozyme (bottom). Supplementary Fig. S7 shows the Ringer profiles and correlation plots for all residues with P CC ≤ 0.95 (ten out of 485 residues).