FIGURE 1.
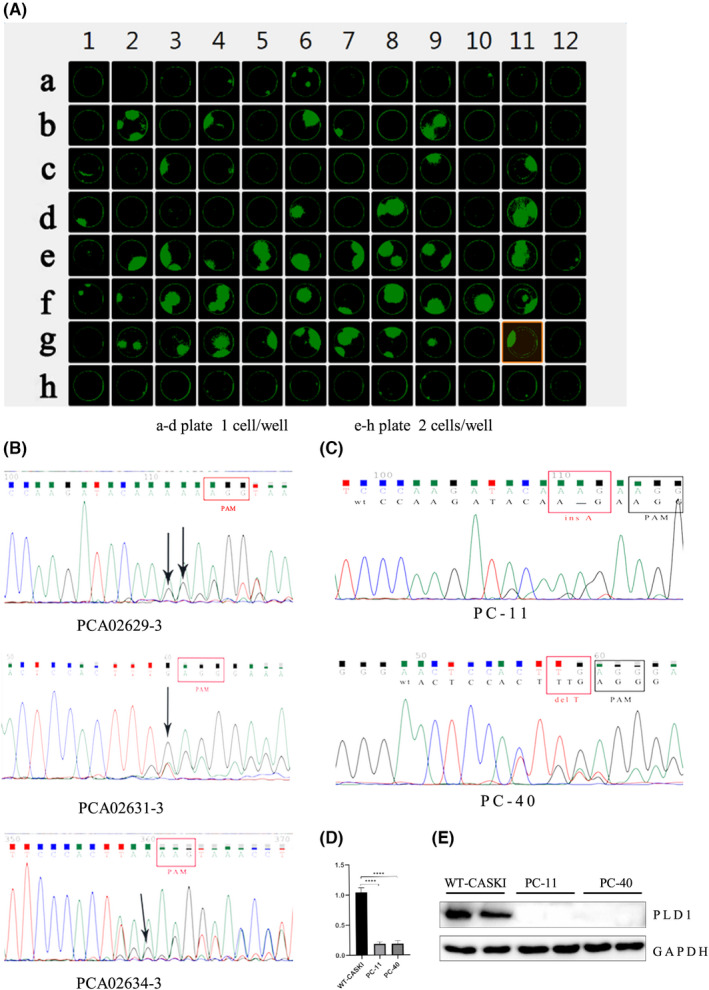
The efficiency of PLD1 knockout by CRISPR/Cas 9. (A) The optimal inoculum density was 2 cells/well for monoclonal culture. (B) Validation of sgRNA cleavage activity through sequence mapping. The three sgRNAs, PCA02629‐3, PCA02631‐3 and PCA02634‐3, showed higher cleavage efficiency. (C) Validation of PLD1 gene mutation in positive cells by Sanger sequencing. The PC‐11 cells carry insertion A base mutation and the PC‐40 cells carry deletion T base mutation (partial sequence). (D) qRT–PCR was used to verify the PLD1 mRNA level (E) Western blot analysis confirmed that the expression of PLD1 was significantly reduced in PC‐11 and PC‐40 cells
