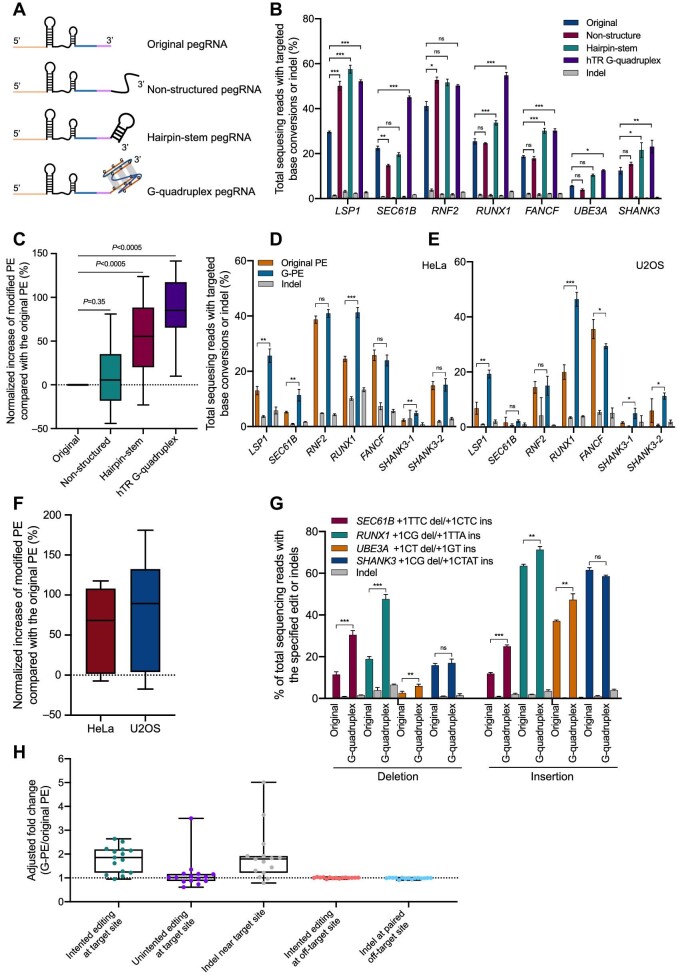
Figure 1.
G-quadruplex-modified pegRNA enhances prime editing efficiency at endogenous target sites. (A) A schematic of pegRNA with 3′ extended modification. An original pegRNA consists of spacer (orange), scaffold (black), RT (blue), and PBS (purple). The structured RNA modification is fused to the 3′ terminal of pegRNA, forming non-structured pegRNA, hairpin-stem pegRNA, and G-quadruplex pegRNA. (B) G-quadruplex-modified pegRNA observably enhances prime editing activity at endogenous target sites in HEK293T cells. PCR amplicons from the target regions were analyzed by targeted deep sequencing. The reads only harboring correct edit were counted to evaluate the editing efficiency, and the reads harboring any unintended insertion or deletion were counted to evaluate the indel frequency. The gray bar indicates the indel frequency coupled with the editing efficiency indicated by the left closest bar. (C) Statistical analysis of normalized increase of targeted base conversions in B. Means from three independent experiments were used for analysis. (D and E) Comparison of the prime editing efficiencies of the original PE and G-PE at endogenous sites in HeLa and U2OS cells. The gray bar indicates the indel frequency coupled with the editing efficiency indicated by the left closest bar. (F) Statistical analysis of normalized increase of targeted base conversions in D and E (HeLa and U2OS cells). Means from three independent experiments were used for analysis. (G) Increasing targeted deletion and insertion efficiency by G-PE in HEK293T cells. The gray bar indicates the indel frequency coupled with the editing efficiency indicated by the left closest bar. (H) The correct editing, unintended editing, and indel frequencies induced by G-PE normalized to those of the original PE at 15 endogenous targets. The off-target editing frequencies of pegRNA and nick sgRNA of G-PE normalized to those of the original PE at three endogenous targets in HEK293T cells. The analysis of adjusted fold increase is described in the Supplementary Materials and methods. Mean ± SD, n = 3 independent experiments (*P < 0.05, **P < 0.005, ***P < 0.0005).