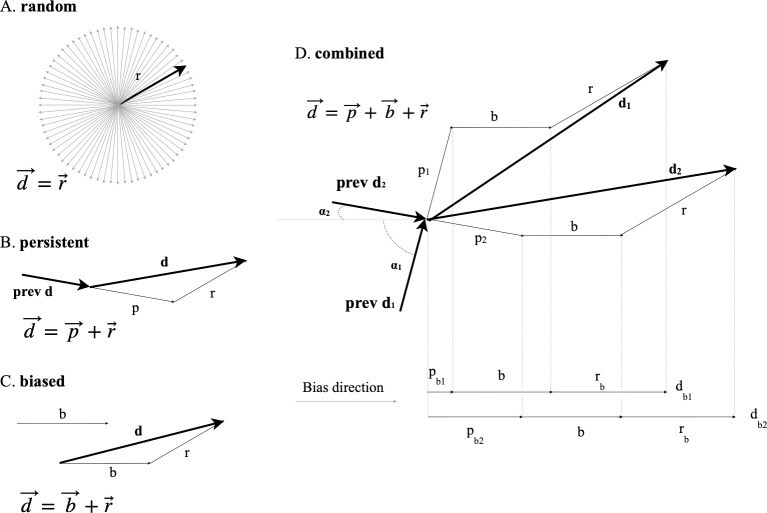
Fig 1. A three component model for single cell movement.
In the case of purely diffusive motion (A), cell displacement is modelled as a random vector of radius r. Persistent and biased movement (B and C), respectively add a persistence (p) or bias (b) vector to the random one. In the general case of combined movement (D), displacements are a combination of the three vectors and the directional component (db) consists of the whole bias module with the addition of random and persistence contributions (respectively rb and pb). The persistence contribution depends on the difference between bias and persistence angle (α angle); its length increases, according to the cosine function, when the α angles decreases.