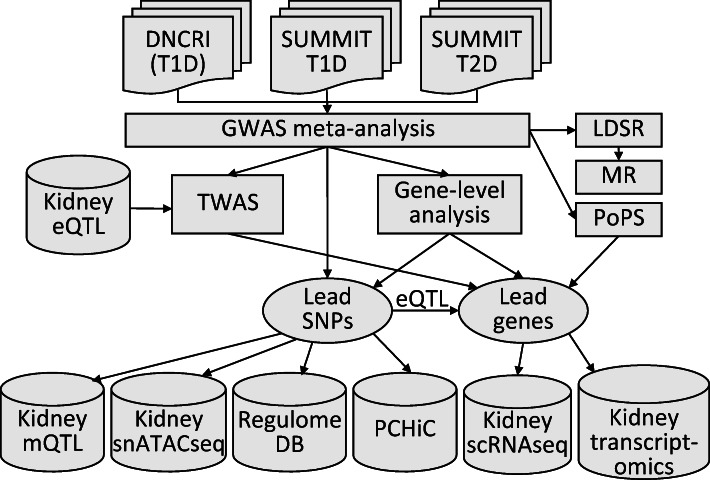
Fig. 1.
Schematic illustration of the study design, from GWAS meta-analysis to integration with various omics data sets. GWAS meta-analysis for ten different phenotypic definitions of DKD included up to 26,785 individuals with either type 1 or type 2 diabetes from the previous DNCRI and SUMMIT GWAS meta-analyses. The TWAS integrated the GWAS meta-analysis results with kidney eQTL data for tubular and glomerular compartments, identifying genes with differential expression in DKD. The mQTL data identified SNPs associated with DNA methylation at CpG sites. Single nucleus Assay for Transposase-Accessible Chromatin using sequencing (snATACseq) was informative of chromatin openness in various kidney cell types. The RegulomeDB is a database with extensive epigenetic annotation for SNPs. The promoter capture HiC (PCHiC) sequencing data identified sequence interaction with gene promoters, proposing target genes. Kidney transcriptomics provided data on gene expression in glomerular and tubular tissue in nephrectomy samples, or in Pima Indian biopsies, correlated with various renal variables. scRNAseq, single-cell RNA sequencing; T1D, type 1 diabetes; T2D, type 2 diabetes