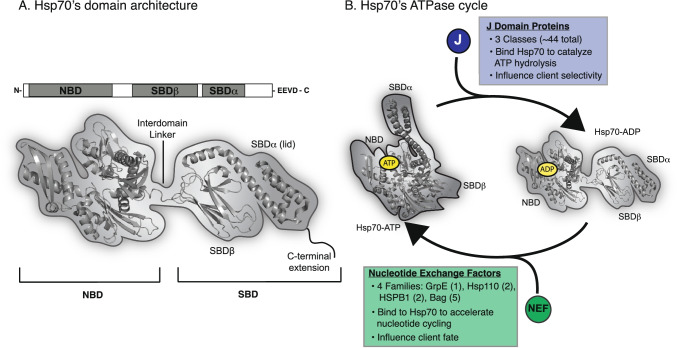
Fig. 1.
Members of the Hsp70 family have a conserved architecture and ATPase cycle. A General structure of Hsp70 family members, including a nucleotide-binding domain (NBD), substrate-binding domain (SBD). The SBD is sub-divided into SBDβ, SBDα (lid) and C-terminal extension. The various Hsp70 orthologs vary in the length and composition of the C-terminal region and cytoplasmic isoforms of eukaryotic Hsp70s also have an EEVD motif. The structure of the prokaryotic Hsp70, DnaK (PDB 2KHO), is shown, along with a cartoon representation. B Schematic of the ATPase cycle of Hsp70s (PDB 2KHO and 5NRO), highlighting the conformational changes that accompany hydrolysis and the roles of the co-chaperones: J-domain proteins (JDPs) and nucleotide exchange factors (NEFs)