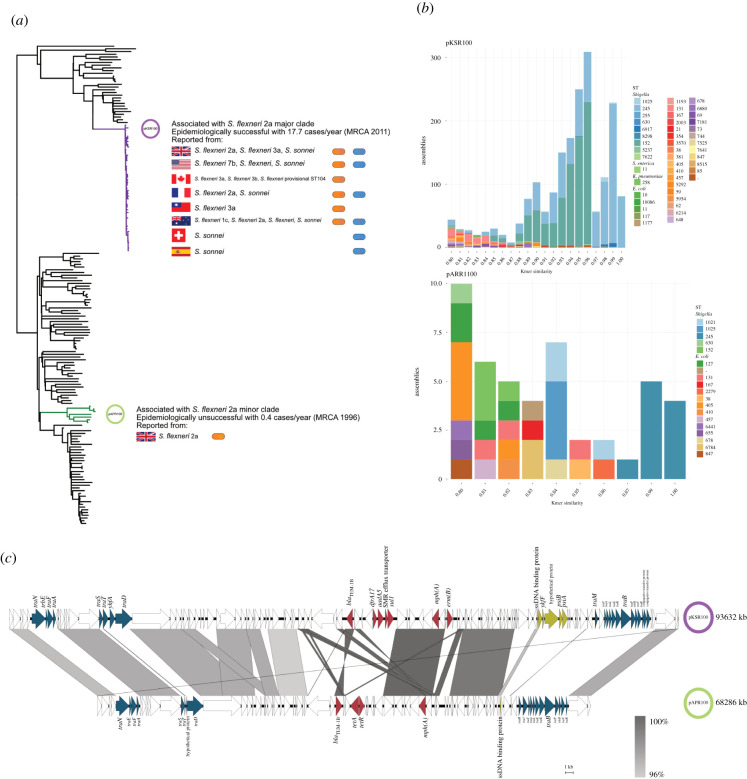
Figure 1.
Relative epidemiological success and comparative genomics of pKSR100 and pAPR100. The disparate epidemiological success of pKSR100- and pAPR100- bearing clades in a cross section of 179 Shigella isolates from UK surveillance data between 2008 and 2014 [26]. The pKSR100-bearing major clade is highlighted in purple and the pARP100-bearing minor clade is highlighted in green (a). The pKSR100 associated major clade had a higher case rate despite a more recent MRCA (most recent common ancestor) compared to the lower case rate and older MRCA of the minor clade, highlighting the more rapid spread of the major clade through the UK. The information alongside the clades depict the disparate global and species distribution of other pKSR100 bacterial hosts being reported in multiple countries across multiple Shigella subtypes (as determined by BLASTn against the NCBI non-redundant database and literature review). pKSR100 and pKSR100-like plasmids have been detected in and reported from eight countries and in multiple Shigella subtypes. By contrast, pAPR100 has only been detected in S. flexneri 2a in the UK. (b) Species and sequence type distribution of genomes sharing kmer similarities of greater than 0.80 with pKSR100 or pAPR100 from across greater than 600 000 publicly available bacterial genomes (in the 661 K COBS data structure). (c) Comparison of the genetic content of both plasmids. Areas of synteny (using a cut-off of 95% BLAST identity) are shown intervening grey bars coloured according to the inlaid legend. Regions with variation between the two plasmids are broadly categorized into three main groups; (i) conjugation machinery related (blue), (ii) SOS response alleviation related (green) and (iii) AMR related (red). (Online version in colour.)