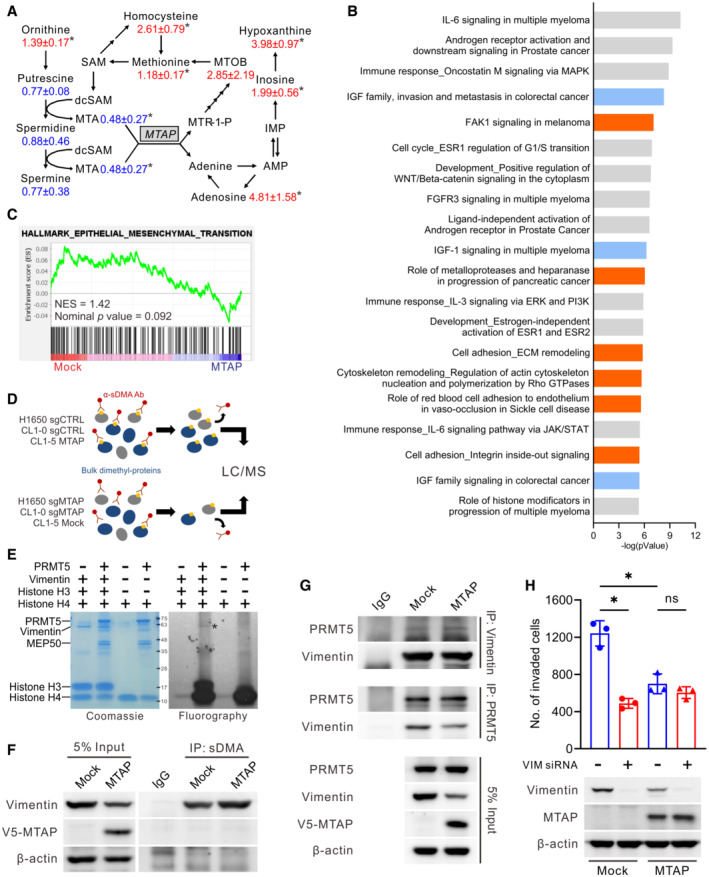
Figure EV2. MTAP‐mediated metabolism, signaling pathways, and methylproteome.
-
AMTAP‐mediated metabolic alterations in polyamine, methionine, and adenine salvage pathways. The numbers are the average fold change intensities and associated errors for metabolites of CL1‐5 MTAP/CL1‐5 Mock (Student t test, n = 3, biological replicates, *P < 0.05).
-
BTop 20 ranking MTAP‐altered pathways and cellular processes identified by the MetaCore analytical suite (version 20.4 build 70,300). Blue bars: IGF‐related pathways; Orange bars: cell adhesion/cytoskeleton remodeling and invasion‐related pathways.
-
CGene Set Enrichment Analysis (GSEA) of expression microarray data from CL1‐5 Mock and MTAP‐overexpressing cells were performed using the gene set of HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION.
-
DSchematic diagram of the identification of differentially symmetrically dimethylated proteins.
-
EIn vitro methylation of vimentin by PRMT5. Tritiated proteins were separated by SDS‐PAGE, stained with Coomassie blue (left), dried and analyzed by fluorography (right). Histone H3 and H4 proteins were used as positive controls. *: tritiated vimentin.
-
FImmunoprecipitation analysis for vimentin dimethylation in CL1‐5 Mock and MTAP‐overexpressing cells. Data shown are representative of three independent experiments.
-
GImmunoprecipitation analysis of the association between endogenous vimentin and endogenous PRMT5 in CL1‐5 Mock and MTAP‐overexpressing cells. Data shown are representative of three independent experiments.
-
HCL1‐5 MTAP‐overexpressing and Mock cells were transfected with vimentin siRNAs for 72 h and analyzed by Boyden chamber invasion assays (mean ± SD, Student t test, n = 3, biological replicates, *P < 0.05). The silence efficiency of vimentin was examined by Western blot.
Source data are available online for this figure.
