-
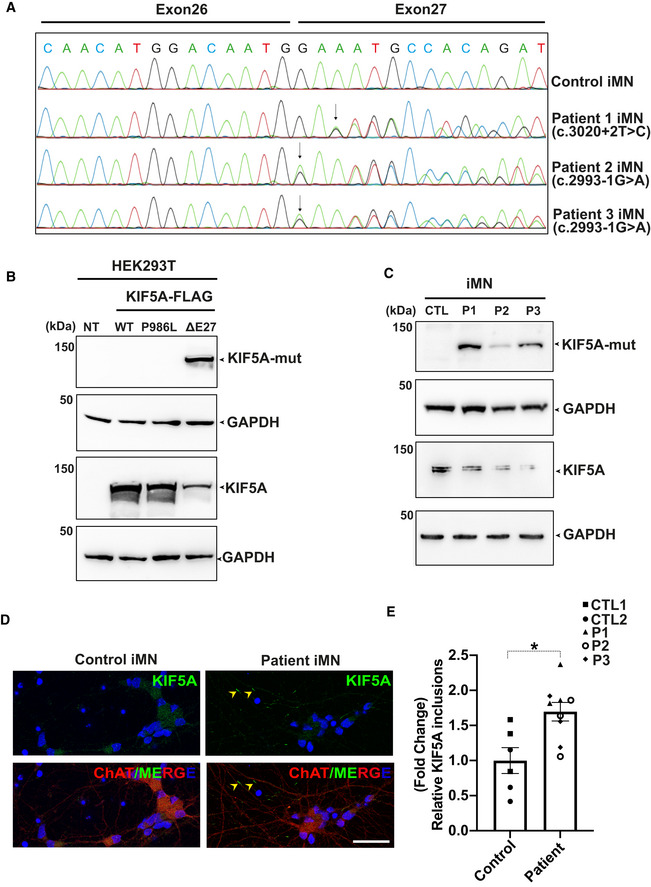
A
Sanger sequencing chromatograms of cDNA from control and patient‐derived iMNs (P1–P3) showing the effect of mutations on KIF5A transcripts. While the c.3020+2T>C mutation causes exon 27 skipping, the c.2993‐1G>A mutation results in an aberrant transcript with a splice acceptor shifted only by a single nucleotide, leading to a frameshift but containing exon 27. The respective protein product predicted for both mutations comprise the same C‐terminal aberrant 39 amino acid end.
-
B
Western blot analysis for validation of the custom mutant‐specific antibody using transfected HEK293T cells. While an antibody against the total KIF5A (lower panel) recognized both wild‐type and mutant proteins, the custom antibody targeting the aberrant “neopeptide” detected ΔExon27 specifically (upper panel). NT: No transfection.
-
C
Mutant KIF5A proteins are endogenously expressed in iMNs, detected by using the custom mutant‐specific antibody directed against the C‐terminal aberrant 39 amino acid sequence. CTL, Control; P1, Patient 1; P2, Patient 2; P3, Patient 3, two independent experiments.
-
D
Representative images of KIF5A distribution (arrowheads) in control and patient iMNs. Scale bar: 20 μm.
-
E
Abundance of KIF5A inclusions after normalizing to total neuron numbers. Graph represents data from n = 146 control 1 (CTL1), n = 134 control 2 (CTL2), n = 154 patient 1 (P1), n = 162 patient 2 (P2) and n = 133 patient 3 (P3) iMNs from three individual experiments. Bars indicate mean ± SD. Statistical analysis was performed using Mann–Whitney Wilcoxon Test (*P < 0.05).